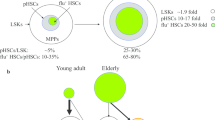
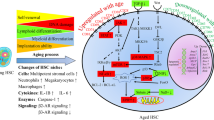
Abstract
Hematopoietic stem cells (HSCs) undergo progressive functional decline over time due to both internal and external stressors, leading to aging of the hematopoietic system. A comprehensive understanding of the molecular mechanisms underlying HSC aging will be valuable in developing novel therapies for HSC rejuvenation and to prevent the onset of several age-associated diseases and hematological malignancies. This review considers the general causes of HSC aging that range from cell-intrinsic factors to cell-extrinsic factors. In particular, epigenetics and inflammation have been implicated in the linkage of HSC aging, clonality, and oncogenesis. The challenges in clarifying mechanisms of HSC aging have accelerated the development of therapeutic interventions to rejuvenate HSCs, the major goal of aging research; these details are also discussed in this review.


Similar content being viewed by others
Data availability
Not applicable.
References
Hayflick L, Moorhead PS (1961) The serial cultivation of human diploid cell strains. Exp Cell Res 25:585–621. https://doi.org/10.1016/0014-4827(61)90192-6
Harley CB, Futcher AB, Greider CW (1990) Telomeres shorten during ageing of human fibroblasts. Nature 345:458–460. https://doi.org/10.1038/246170a0
Kenyon CJ (2010) The genetics of ageing. Nature 464:504–512. https://doi.org/10.1038/nature08980
Buffenstein R (2005) The naked mole-rat : a new long-living model. J Gerontol A Biol Sci Med Sci 60:1369–1377
Gordon LB, Rothman FG, López-Otín C, Misteli T (2014) Progeria: a paradigm for translational medicine. Cell 156:400–407. https://doi.org/10.1016/j.cell.2013.12.028
Genovese G, Kähler AK, Handsaker RE et al (2014) Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med 371:2477–2487. https://doi.org/10.1056/NEJMoa1409405
Jaiswal S, Fontanillas P, Flannick J et al (2014) Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med 371:2488–2498. https://doi.org/10.1056/NEJMoa1408617
Xie M, Lu C, Wang J et al (2014) Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med 20:1472–1478. https://doi.org/10.1038/nm.3733
Jaiswal S, Natarajan P, Silver AJ et al (2017) Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N Engl J Med 377:111–121. https://doi.org/10.1056/NEJMoa1701719
Vanasse GJ, Berliner N (2010) Anemia in elderly patients: an emerging problem for the 21st century. Hematol Am Soc Hematol Educ Progr. https://doi.org/10.1182/asheducation-2010.1.271
Guralnik JM, Eisenstaedt RS, Ferrucci L et al (2004) Prevalence of anemia in persons 65 years and older in the United States: evidence for a high rate of unexplained anemia. Blood 104:2263–2268. https://doi.org/10.1182/blood-2004-05-1812
Malcovati L, Gallì A, Travaglino E et al (2017) Clinical significance of somatic mutation in unexplained blood cytopenia. Blood 129:3371–3378. https://doi.org/10.1182/blood-2017-01-763425
Kwok B, Hall JM, Witte JS et al (2015) MDS-associated somatic mutations and clonal hematopoiesis are common in idiopathic cytopenias of undetermined significance. Blood 126:2355–2361. https://doi.org/10.1182/blood-2015-08-667063
World health organization (2020) The top 10 causes of death
Denkinger MD, Leins H, Schirmbeck R et al (2015) HSC aging and senescent immune remodeling. Trends Immunol 36:815–824. https://doi.org/10.1016/j.it.2015.10.008
Sudo K, Ema H, Morita Y, Nakauchi H (2000) Age-associated characteristics of murine hematopoietic stem cells. J Exp Med 192:1273–1280. https://doi.org/10.1084/jem.192.9.1273
Rossi DJ, Bryder D, Zahn JM et al (2005) Cell intrinsic alterations underlie hematopoietic stem cell aging. Proc Natl Acad Sci USA 102:9194–9199. https://doi.org/10.1073/pnas.0503280102
Cho RH, Sieburg HB, Muller-Sieburg CE (2008) A new mechanism for the aging of hematopoietic stem cells: aging changes the clonal composition of the stem cell compartment but not individual stem cells. Blood 111:5553–5561. https://doi.org/10.1182/blood-2007-11-123547
Bernitz JM, Kim HS, MacArthur B et al (2016) Hematopoietic stem cells count and remember self-renewal divisions. Cell 167:1296-1309.e10. https://doi.org/10.1016/j.cell.2016.10.022
Yamamoto R, Wilkinson AC, Ooehara J et al (2018) Large-scale clonal analysis resolves aging of the mouse hematopoietic stem cell compartment. Cell Stem Cell 22:600-607.e4. https://doi.org/10.1016/j.stem.2018.03.013
Naylor K, Li G, Vallejo AN et al (2005) The influence of age on T cell generation and TCR diversity. J Immunol 174:7446–7452. https://doi.org/10.4049/jimmunol.174.11.7446
Qi Q, Liu Y, Cheng Y et al (2014) Diversity and clonal selection in the human T-cell repertoire. Proc Natl Acad Sci USA 111:13139–13144. https://doi.org/10.1073/pnas.1409155111
Miller RA (1996) The aging immune system: primer and prospectus. Science (80-) 273:70–74. https://doi.org/10.1126/science.273.5271.70
Chae WJ, Bothwell ALM (2018) Canonical and non-canonical wnt signaling in immune cells. Trends Immunol 39:830–847. https://doi.org/10.1016/j.it.2018.08.006
Florian MC, Nattamai KJ, Dörr K et al (2013) A canonical to non-canonical Wnt signalling switch in haematopoietic stem-cell ageing. Nature 503:392–396. https://doi.org/10.1038/nature12631
Florian MC, Dörr K, Niebel A et al (2012) Cdc42 activity regulates hematopoietic stem cell aging and rejuvenation. Cell Stem Cell 10:520–530. https://doi.org/10.1016/j.stem.2012.04.007
Oh J, Lee YD, Wagers AJ (2014) Stem cell aging: mechanisms, regulators and therapeutic opportunities. Nat Med 20:870–880. https://doi.org/10.1038/nm.3651
Akunuru S, Geiger H (2016) Aging, clonality, and rejuvenation of hematopoietic stem cells. Trends Mol Med 22:701–712. https://doi.org/10.1016/j.molmed.2016.06.003
De Haan G, Lazare SS (2018) Aging of hematopoietic stem cells. Blood 131:479–487. https://doi.org/10.1182/blood-2017-06-746412
Orkin SH, Zon LI (2008) Hematopoiesis: an evolving paradigm for stem cell biology. Cell 132:631–644
Dykstra B, Olthof S, Schreuder J et al (2011) Clonal analysis reveals multiple functional defects of aged murine hematopoietic stem cells. J Exp Med 208:2691–2703. https://doi.org/10.1084/jem.20111490
Liang Y, Van ZG, Szilvassy SJ (2005) Effects of aging on the homing and engraftment of murine hematopoietic stem and progenitor cells. Blood 106:1479–1487. https://doi.org/10.1182/blood-2004-11-4282
Cheng T, Rodrigues N, Shen H et al (2000) Hematopoietic stem cell quiescence maintained by p21(cip1/waf1). Science (80-) 287:1804–1809. https://doi.org/10.1126/science.287.5459.1804
He S, Sharpless NE (2017) Senescence in Health and Disease. Cell 169:1000–1011. https://doi.org/10.1016/j.cell.2017.05.015
Dimri GP, Lee X, Basile G et al (1995) A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA 92:9363–9367. https://doi.org/10.1073/pnas.92.20.9363
Melk A, Schmidt BMW, Takeuchi O et al (2004) Expression of p16INK4a and other cell cycle regulator and senescence associated genes in aging human kidney. Kidney Int 65:510–520. https://doi.org/10.1111/j.1523-1755.2004.00438.x
Molofsky AV, Slutsky SG, Joseph NM et al (2006) Increasing p16INK4a expression decreases forebrain progenitors and neurogenesis during ageing. Nature 443:448–452. https://doi.org/10.1038/nature05091
Sousa-Victor P, Gutarra S, García-Prat L et al (2014) Geriatric muscle stem cells switch reversible quiescence into senescence. Nature 506:316–321. https://doi.org/10.1038/nature13013
Janzen V, Forkert R, Fleming HE et al (2006) Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16 INK4a. Nature 443:421–426. https://doi.org/10.1038/nature05159
Krishnamurthy J, Torrice C, Ramsey MR et al (2004) Ink4a/Arf expression is a biomarker of aging. J Clin Invest 114:1299–1307. https://doi.org/10.1172/JCI22475
Signer RAJ, Montecino-Rodriguez E, Witte ON, Dorshkind K (2008) Aging and cancer resistance in lymphoid progenitors are linked processes conferred by p16Ink4a and Arf. Genes Dev 22:3115–3120. https://doi.org/10.1101/gad.1715808
Liu Y, Johnson SM, Fedoriw Y et al (2011) Expression of p16INK4a prevents cancer and promotes aging in lymphocytes. Blood 117:3257–3267. https://doi.org/10.1182/blood-2010-09-304402
Zhu J, Woods D, McMahon M, Bishop JM (1998) Senescence of human fibroblasts induced by oncogenic Raf. Genes Dev 12:2997–3007. https://doi.org/10.1101/gad.12.19.2997
Lin AW, Barradas M, Stone JC et al (1998) Premature senescence involving p53 and p16 is activated in response to constitutive MEK/MAPK mitogenic signaling. Genes Dev 12:3008–3019. https://doi.org/10.1101/gad.12.19.3008
Serrano M, Lin AW, McCurrach ME et al (1997) Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16(INK4a). Cell 88:593–602. https://doi.org/10.1016/S0092-8674(00)81902-9
Chicas A, Wang X, Zhang C et al (2010) Dissecting the unique role of the retinoblastoma tumor suppressor during cellular senescence. Cancer Cell 17:376–387. https://doi.org/10.1016/j.ccr.2010.01.023
Rodier F, Coppé JP, Patil CK et al (2009) Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat Cell Biol 11:973–979. https://doi.org/10.1038/ncb1909
Coppé JP, Patil CK, Rodier F et al (2008) Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. https://doi.org/10.1371/journal.pbio.0060301
Johmura Y, Shimada M, Misaki T et al (2014) Necessary and sufficient role for a mitosis skip in senescence induction. Mol Cell 55:73–84. https://doi.org/10.1016/j.molcel.2014.05.003
Zindy F, Quelle DE, Roussel MF, Sherr CJ (1997) Expression of the p16INK4a tumor suppressor versus other INK4 family members during mouse development and aging. Oncogene 15:203–211. https://doi.org/10.1038/sj.onc.1201178
Sharpless NE, Bardeesy N, Lee KH et al (2001) Loss of p16Ink4a with retention of p19 predisposes mice to tumorigenesis. Nature 413:86–91. https://doi.org/10.1038/35092592
Lujambio A, Akkari L, Simon J et al (2013) Non-cell-autonomous tumor suppression by p53. Cell 153:449–460. https://doi.org/10.1016/j.cell.2013.03.020
Xue W, Zender L, Miething C et al (2007) Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 445:656–660. https://doi.org/10.1038/nature05529
Krizhanovsky V, Yon M, Dickins RA et al (2008) Senescence of activated stellate cells limits liver fibrosis. Cell 134:657–667. https://doi.org/10.1016/j.cell.2008.06.049
Kang TW, Yevsa T, Woller N et al (2011) Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature 479:547–551. https://doi.org/10.1038/nature10599
Krtolica A, Parrinello S, Lockett S et al (2001) Senescent fibroblasts promote epithelial cell growth and tumorigenesis: a link between cancer and aging. Proc Natl Acad Sci USA 98:12072–12077. https://doi.org/10.1073/pnas.211053698
Ohanna M, Giuliano S, Bonet C et al (2011) Senescent cells develop a parp-1 and nuclear factor-κB-associated secretome (PNAS). Genes Dev 25:1245–1261. https://doi.org/10.1101/gad.625811
Coppe JP, Kauser K, Campisi J, Beauséjour CM (2006) Secretion of vascular endothelial growth factor by primary human fibroblasts at senescence. J Biol Chem 281:29568–29574. https://doi.org/10.1074/jbc.M603307200
Lasry A, Ben-Neriah Y (2015) Senescence-associated inflammatory responses: aging and cancer perspectives. Trends Immunol 36:217–228. https://doi.org/10.1016/j.it.2015.02.009
Michaloglou C, Vredeveld LCW, Soengas MS et al (2005) BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature 436:720–724. https://doi.org/10.1038/nature03890
Braig M, Lee S, Loddenkemper C et al (2005) Oncogene-induced senescence as an initial barrier in lymphoma development. Nature 436:660–665. https://doi.org/10.1038/nature03841
Chen Z, Trotman LC, Shaffer D et al (2005) Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature 436:725–730. https://doi.org/10.1038/nature03918
Biavasco R, Lettera E, Giannetti K et al (2021) Oncogene-induced senescence in hematopoietic progenitors features myeloid restricted hematopoiesis, chronic inflammation and histiocytosis. Nat Commun 12:1–18. https://doi.org/10.1038/s41467-021-24876-1
Attema JL, Pronk CJH, Norddahl GL et al (2009) Hematopoietic stem cell ageing is uncoupled from p16 INK4A -mediated senescence. Oncogene 28:2238–2243. https://doi.org/10.1038/onc.2009.94
Sun D, Luo M, Jeong M et al (2014) Epigenomic profiling of young and aged HSCs reveals concerted changes during aging that reinforce self-renewal. Cell Stem Cell 14:673–688. https://doi.org/10.1016/j.stem.2014.03.002
Rogakou EP, Pilch DR, Orr AH et al (1998) DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 273:5858–5868. https://doi.org/10.1074/jbc.273.10.5858
Singh NP, McCoy MT, Tice RR, Schneider EL (1988) A simple technique for quantitation of low levels of DNA damage in individual cells. Exp Cell Res 175:184–191. https://doi.org/10.1016/0014-4827(88)90265-0
Beerman I, Seita J, Inlay MA et al (2014) Quiescent hematopoietic stem cells accumulate DNA damage during aging that is repaired upon entry into cell cycle. Cell Stem Cell 15:37–50. https://doi.org/10.1016/j.stem.2014.04.016
Rossi DJ, BryderSeita DJ et al (2007) Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature 447:725–729. https://doi.org/10.1038/nature05862
Moehrle BM, Nattamai K, Brown A et al (2015) Stem cell-specific mechanisms ensure genomic fidelity within HSCs and upon aging of HSCs. Cell Rep 13:2412–2424. https://doi.org/10.1016/j.celrep.2015.11.030
Mohrin M, Bourke E, Alexander D et al (2010) Hematopoietic stem cell quiescence promotes error-prone DNA repair and mutagenesis. Cell Stem Cell 7:174–185. https://doi.org/10.1016/j.stem.2010.06.014
Insinga A, Cicalese A, Faretta M et al (2013) DNA damage in stem cells activates p21, inhibits p53, and induces symmetric self-renewing divisions. Proc Natl Acad Sci USA 110:3931–3936. https://doi.org/10.1073/pnas.1213394110
Burtner CR, Kennedy BK (2010) Progeria syndromes and ageing: what is the connection? Nat Rev Mol Cell Biol 11:567–578. https://doi.org/10.1038/nrm2944
Ito K, Hirao A, Arai F et al (2004) Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature 431:997–1002. https://doi.org/10.1038/nature02989
Nijnik A, Woodbine L, Marchetti C et al (2007) DNA repair is limiting for haematopoietic stem cells during ageing. Nature 447:686–690. https://doi.org/10.1038/nature05875
Prasher JM, Lalai AS, Heijmans-Antonissen C et al (2005) Reduced hematopoietic reserves in DNA interstrand crosslink repair-deficient Ercc1-/- mice. EMBO J 24:861–871. https://doi.org/10.1038/sj.emboj.7600542
Reese JS, Liu L, Gerson SL (2003) Repopulating defect of mismatch repair-deficient hematopoietic stem cells. Blood 102:1626–1633. https://doi.org/10.1182/blood-2002-10-3035
Zhang QS, Marquez-Loza L, Eaton L et al (2010) Fancd2-/- mice have hematopoietic defects that can be partially corrected by resveratrol. Blood 116:5140–5148. https://doi.org/10.1182/blood-2010-04-278226
Ruzankina Y, Pinzon-Guzman C, Asare A et al (2007) Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell 1:113–126. https://doi.org/10.1016/j.stem.2007.03.002
Santos MA, Faryabi RB, Ergen AV et al (2014) DNA-damage-induced differentiation of leukaemic cells as an anti-cancer barrier. Nature 514:107–111. https://doi.org/10.1038/nature13483
Halazonetis TD, Gorgoulis VG, Bartek J (2008) An oncogene-induced DNA damage model for cancer development. Science (80-) 319:1352–1355. https://doi.org/10.1126/science.1140735
Abbas HA, MacCio DR, Coskun S et al (2010) Mdm2 Is required for survival of hematopoietic stem cells/progenitors via dampening of ROS-induced p53 activity. Cell Stem Cell 7:606–617. https://doi.org/10.1016/j.stem.2010.09.013
Shay JW, Wright WE (2019) Telomeres and telomerase: three decades of progress. Nat Rev Genet 20:299–309. https://doi.org/10.1038/s41576-019-0099-1
Vaziri H, Dragowska W, Allsopp RC et al (1994) Evidence for a mitotic clock in human hematopoietic stem cells: Loss of telomeric DNA with age. Proc Natl Acad Sci USA 91:9857–9860. https://doi.org/10.1073/pnas.91.21.9857
Allsopp RC, Morin GB, DePinho R et al (2003) Telomerase is required to slow telomere shortening and extend replicative lifespan of HSCs during serial transplantation. Blood 102:517–520. https://doi.org/10.1182/blood-2002-07-2334
Allsoppi RC, Morin GB, Horner JW et al (2003) Effect of TERT over-expression on the long-term transplantation capacity of hematopoietic stem cells [2]. Nat Med 9:369–371. https://doi.org/10.1038/nm0403-369
Marrone A, Stevens D, Vulliamy T et al (2004) Heterozygous telomerase RNA mutations found in dyskeratosis congenita and aplastic anemia reduce telomerase activity via haploinsufficiency. Blood 104:3936–3942. https://doi.org/10.1182/blood-2004-05-1829
Yamaguchi H, Calado RT, Ly H et al (2005) Mutations in TERT, the gene for telomerase reverse transcriptase, in aplastic anemia. N Engl J Med 352:1413–1424. https://doi.org/10.1056/nejmoa042980
Vulliamy T, Marrone A, Szydlo R et al (2004) Disease anticipation is associated with progressive telomere shortening in families with dyskeratosis congenita due to mutations in TERC. Nat Genet 36:447–449. https://doi.org/10.1038/ng1346
Vulliamy T, Marrone A, Goldman F et al (2001) The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature 413:432–435. https://doi.org/10.1038/35096585
Mitchell JR, Wood E, Collins K (1999) A telomerase component is defective in the human disease dyskeratosis congenita. Nature 402:551–555. https://doi.org/10.1038/990141
Greenberg MVC, Bourc’his D (2019) The diverse roles of DNA methylation in mammalian development and disease. Nat Rev Mol Cell Biol 20:590–607. https://doi.org/10.1038/s41580-019-0159-6
Trowbridge JJ, Snow JW, Kim J, Orkin SH (2009) DNA methyltransferase 1 Is essential for and uniquely regulates hematopoietic stem and progenitor cells. Cell Stem Cell 5:442–449. https://doi.org/10.1016/j.stem.2009.08.016
Challen GA, Sun D, Jeong M et al (2012) Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet 44:23–31. https://doi.org/10.1038/ng.1009
Moran-Crusio K, Reavie L, Shih A et al (2011) Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell 20:11–24. https://doi.org/10.1016/j.ccr.2011.06.001
Quivoron C, Couronné L, Della Valle V et al (2011) TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell 20:25–38. https://doi.org/10.1016/j.ccr.2011.06.003
Adelman ER, Huang HT, Roisman A et al (2019) Aging human hematopoietic stem cells manifest profound epigenetic reprogramming of enhancers that may predispose to leukemia. Cancer Discov 9:1080–1101. https://doi.org/10.1158/2159-8290.CD-18-1474
Schuettengruber B, Bourbon HM, Di Croce L, Cavalli G (2017) Genome regulation by polycomb and trithorax: 70 years and counting. Cell 171:34–57. https://doi.org/10.1016/j.cell.2017.08.002
Beerman I, Rossi DJ (2015) Epigenetic control of stem cell potential during homeostasis, aging, and disease. Cell Stem Cell 16:613–625. https://doi.org/10.1016/j.stem.2015.05.009
Jude CD, Climer L, Xu D et al (2007) Unique and independent roles for MLL in adult hematopoietic stem cells and progenitors. Cell Stem Cell 1:324–337. https://doi.org/10.1016/j.stem.2007.05.019
Xie H, Xu J, Hsu JH et al (2014) Polycomb repressive complex 2 regulates normal hematopoietic stem cell function in a developmental-stage-specific manner. Cell Stem Cell 14:68–80. https://doi.org/10.1016/j.stem.2013.10.001
Park IK, Qian D, Kiel M et al (2003) Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature 423:302–305. https://doi.org/10.1038/nature01587
McMahon KA, Hiew SYL, Hadjur S et al (2007) Mll Has a critical role in fetal and adult hematopoietic stem cell self-renewal. Cell Stem Cell 1:338–345. https://doi.org/10.1016/j.stem.2007.07.002
Bird A (2002) DNA methylation patterns and epigenetic memory. Genes Dev 16:6–21. https://doi.org/10.1101/gad.947102
Fraga MF, Ballestar E, Paz MF et al (2005) Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci USA 102:10604–10609
Yu VWC, Yusuf RZ, Oki T et al (2016) Epigenetic memory underlies cell-autonomous heterogeneous behavior of hematopoietic stem cells. Cell 167:1310-1322.e17. https://doi.org/10.1016/j.cell.2016.10.045
Cheung P, Vallania F, Warsinske HC et al (2018) Single-cell chromatin modification profiling reveals increased epigenetic variations with aging. Cell 173:1385-1397.e14. https://doi.org/10.1016/j.cell.2018.03.079
Bigarella CL, Liang R, Ghaffari S (2014) Stem cells and the impact of ROS signaling. Development 141:4206–4218. https://doi.org/10.1242/dev.107086
Harman D (1956) Aging: a theory based on free radical and radiation chemistry. J Gerontol 11:298–300. https://doi.org/10.1093/geronj/11.3.298
Jang Y, Sharkis SJ (2007) A low level of reactive oxygen species selects for primitive hematopoietic stem cells that may reside in the low-oxygenic niche. Blood 110:3056–3063. https://doi.org/10.1182/blood-2007-05-087759.An
Yahata T, Takanashi T, Muguruma Y et al (2011) Accumulation of oxidative DNA damage restricts the self-renewal capacity of human hematopoietic stem cells. Blood 118:2941–2950. https://doi.org/10.1182/blood-2011-01-330050
Webb AE, Brunet A (2014) FOXO transcription factors: key regulators of cellular quality control. Trends Biochem Sci 39:159–169. https://doi.org/10.1016/j.tibs.2014.02.003
Tothova Z, Kollipara R, Huntly BJ et al (2007) FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell 128:325–339. https://doi.org/10.1016/j.cell.2007.01.003
Miyamoto K, Araki KY, Naka K et al (2007) Foxo3a is essential for maintenance of the hematopoietic stem cell pool. Cell Stem Cell 1:101–112. https://doi.org/10.1016/j.stem.2007.02.001
Wang Y, Hekimi S (2015) Mitochondrial dysfunction and longevity in animals: untangling the knot. Science (80-) 350:1204–1207. https://doi.org/10.1126/science.aac4357
Norddahl GL, Pronk CJ, Wahlestedt M et al (2011) Accumulating mitochondrial DNA mutations drive premature hematopoietic aging phenotypes distinct from physiological stem cell aging. Cell Stem Cell 8:499–510. https://doi.org/10.1016/j.stem.2011.03.009
Ansó E, Weinberg SE, Diebold LP et al (2017) The mitochondrial respiratory chain is essential for haematopoietic stem cell function. Nat Cell Biol 19:614–625. https://doi.org/10.1038/ncb3529
Corral-Debrinski M, Horton T, Lott MT et al (1992) Mitochondrial DNA deletions in human brain: regional variability and increase with advanced age. Nat Genet 2:324–329. https://doi.org/10.1038/ng1292-324
Cortopassi GA, Arnheim N (1990) Detection of a specific mitochondrial DNA deletion in tissues of older humans. Nucleic Acids Res 18:6927–6933. https://doi.org/10.1093/nar/18.23.6927
Soong NW, Hinton DR, Cortopassi G, Arnheim N (1992) Mosaicism for a specific somatic mitochondrial DNA mutation in adult human brain. Nat Genet 2:318–323. https://doi.org/10.1038/ng1292-318
Pikó L, Hougham AJ, Bulpitt KJ (1988) Studies of sequence heterogeneity of mitochondrial DNA from rat and mouse tissues: evidence for an increased frequency of deletions/additions with aging. Mech Ageing Dev 43:279–293. https://doi.org/10.1016/0047-6374(88)90037-1
Harper ME, Monemdjou S, Ramsey JJ, Weindruch R (1998) Age-related increase in mitochondrial proton leak and decrease in ATP turnover reactions in mouse hepatocytes. Am J Physiol-Endocrinol Metab 275:197–206. https://doi.org/10.1152/ajpendo.1998.275.2.e197
Mortensen M, Soilleux EJ, Djordjevic G et al (2011) The autophagy protein Atg7 is essential for hematopoietic stem cell maintenance. J Exp Med 208:455–467. https://doi.org/10.1084/jem.20101145
Ho TT, Warr MR, Adelman ER et al (2017) Autophagy maintains the metabolism and function of young and old stem cells. Nature 543:205–210
Jung HE, Shim YR, Oh JE et al (2019) The autophagy protein Atg5 plays a crucial role in the maintenance and reconstitution ability of hematopoietic stem cells. Immune Netw 19:1–13. https://doi.org/10.4110/in.2019.19.e12
Dong S, Wang Q, Kao YR et al (2021) Chaperone-mediated autophagy sustains haematopoietic stem-cell function. Nature 591:117–123. https://doi.org/10.1038/s41586-020-03129-z
Mohrin M, Shin J, Liu Y et al (2015) A mitochondrial UPR-mediated metabolic checkpoint regulates hematopoietic stem cell aging. Science (80-) 347:1374–1377. https://doi.org/10.1126/science.aaa2361
Jin G, Xu C, Zhang X et al (2018) Atad3a suppresses Pink1-dependent mitophagy to maintain homeostasis of hematopoietic progenitor cells article. Nat Immunol 19:29–40. https://doi.org/10.1038/s41590-017-0002-1
Simsek T, Kocabas F, Zheng J et al (2010) The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. Cell Stem Cell 7:380–390. https://doi.org/10.1016/j.stem.2010.07.011
Takubo K, Nagamatsu G, Kobayashi CI et al (2013) Regulation of glycolysis by Pdk functions as a metabolic checkpoint for cell cycle quiescence in hematopoietic stem cells. Cell Stem Cell 12:49–61. https://doi.org/10.1016/j.stem.2012.10.011
López-Otín C, Galluzzi L, Freije JMP et al (2016) Metabolic control of longevity. Cell 166:802–821. https://doi.org/10.1016/j.cell.2016.07.031
Ito K, Suda T (2014) Metabolic requirements for the maintenance of self-renewing stem cells. Nat Rev Mol Cell Biol 15:243–256. https://doi.org/10.1038/nrm3772
Kharas MG, Okabe R, Ganis JJ et al (2010) Constitutively active AKT depletes hematopoietic stem cells and induces leukemia in mice. Blood 115:1406–1415. https://doi.org/10.1182/blood-2009-06-229443
Zhang J, Grindley JC, Yin T et al (2006) PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature 441:518–522. https://doi.org/10.1038/nature04747
Yilmaz ÖH, Valdez R, Theisen BK et al (2006) Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature 441:475–482. https://doi.org/10.1038/nature04703
Gan B, Sahin E, Jiang S et al (2008) mTORC1-dependent and -independent regulation of stem cell renewal, differentiation, and mobilization. Proc Natl Acad Sci USA 105:19384–19389. https://doi.org/10.1073/pnas.0810584105
Juntilla MM, Patil VD, Calamito M et al (2010) AKT1 and AKT2 maintain hematopoietic stem cell function by regulating reactive oxygen species. Blood 115:4030–4038. https://doi.org/10.1182/blood-2009-09-241000
Gan B, Hu J, Jiang S et al (2010) Lkb1 regulates quiescence and metabolic homeostasis of haematopoietic stem cells. Nature 468:701–704. https://doi.org/10.1038/nature09595
Chen C, Liu Y, Liu R et al (2008) TSC–mTOR maintains quiescence and function of hematopoietic stem cells by repressing mitochondrial biogenesis and reactive oxygen species. J Exp Med 205:2397–2408. https://doi.org/10.1084/jem.20081297
Kalaitzidis D, Sykes SM, Wang Z et al (2012) MTOR complex 1 plays critical roles in hematopoiesis and pten-loss-evoked leukemogenesis. Cell Stem Cell 11:429–439. https://doi.org/10.1016/j.stem.2012.06.009
Aman Y, Schmauck-Medina T, Hansen M et al (2021) Autophagy in healthy aging and disease. Nat Aging 1:634–650. https://doi.org/10.1038/s43587-021-00098-4
Hidalgo San Jose L, Sunshine MJ, Dillingham CH et al (2020) Modest declines in proteome quality impair hematopoietic stem cell self-renewal. Cell Rep 30:69-80.e6. https://doi.org/10.1016/j.celrep.2019.12.003
Signer RAJ, Magee JA, Salic A, Morrison SJ (2014) Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature 508:49–54. https://doi.org/10.1038/nature13035
Warr MR, Binnewies M, Flach J et al (2013) FOXO3A directs a protective autophagy program in haematopoietic stem cells. Nature 494:323–327. https://doi.org/10.1038/nature11895
Kaushik S, Cuervo AM (2018) The coming of age of chaperone-mediated autophagy. Nat Rev Mol Cell Biol 19:365–381. https://doi.org/10.1038/s41580-018-0001-6
Houtkooper RH, Pirinen E, Auwerx J (2012) Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol 13:225–238. https://doi.org/10.1038/nrm3293
Rimmelé P, Bigarella CL, Liang R et al (2014) Aging-like phenotype and defective lineage specification in SIRT1-deleted hematopoietic stem and progenitor cells. Stem Cell Reports 3:44–59. https://doi.org/10.1016/j.stemcr.2014.04.015
Brown K, Xie S, Qiu X et al (2013) SIRT3 reverses aging-associated degeneration. Cell Rep 3:319–327. https://doi.org/10.1016/j.celrep.2013.01.005
Wang H, Diao D, Shi Z et al (2016) SIRT6 controls hematopoietic stem cell homeostasis through epigenetic regulation of Wnt signaling. Cell Stem Cell 18:495–507. https://doi.org/10.1016/j.stem.2016.03.005
Luo H, Mu WC, Karki R et al (2019) Mitochondrial stress-initiated aberrant activation of the NLRP3 inflammasome regulates the functional deterioration of hematopoietic stem cell aging. Cell Rep 26:945-954.e4. https://doi.org/10.1016/j.celrep.2018.12.101
Grigoryan A, Guidi N, Senger K et al (2018) LaminA/C regulates epigenetic and chromatin architecture changes upon aging of hematopoietic stem cells. Genome Biol 19:1–21. https://doi.org/10.1186/s13059-018-1557-3
Schofield R (1978) The relationship between the spleen colony-forming cell and the haemopoietic stem cell. Blood Cells 4:7–25
Pinho S, Frenette PS (2019) Haematopoietic stem cell activity and interactions with the niche. Nat Rev Mol Cell Biol 20:303–320. https://doi.org/10.1038/s41580-019-0103-9
Gong JK (1978) Endosteal marrow: a rich source of hematopoietic stem cells. Science 199:1443–1445. https://doi.org/10.1126/science.75570
Nilsson SK, Johnston HM, Coverdale JA (2001) Spatial localization of transplanted hemopoietic stem cells: inferences for the localization of stem cell niches. Blood 97:2293–2299. https://doi.org/10.1182/blood.V97.8.2293
Calvi LM, Adams GB, Weibrecht KW et al (2003) Osteoblastic cells regulate the haematopoietic stem cell niche. Nature 425:841–846. https://doi.org/10.1038/nature02040
Zhang J, Niu C, Ye L et al (2003) Identification of the haematopoietic stem cell niche and control of the niche size. Nature 425:836–841. https://doi.org/10.1038/nature02041
Xie Y, Yin T, Wiegraebe W et al (2009) Detection of functional haematopoietic stem cell niche using real-time imaging. Nature 457:97–101. https://doi.org/10.1038/nature07639
Lo Celso C, Fleming HE, Wu JW et al (2009) Live-animal tracking of individual haematopoietic stem/progenitor cells in their niche. Nature 457:92–96. https://doi.org/10.1038/nature07434
Ding L, Morrison SJ (2013) Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches. Nature 495:231–235. https://doi.org/10.1038/nature11885
Katayama Y, Battista M, Kao WM et al (2006) Signals from the sympathetic nervous system regulate hematopoietic stem cell egress from bone marrow. Cell 124:407–421. https://doi.org/10.1016/j.cell.2005.10.041
Méndez-Ferrer S, Lucas D, Battista M, Frenette PS (2008) Haematopoietic stem cell release is regulated by circadian oscillations. Nature 452:442–447. https://doi.org/10.1038/nature06685
Spiegel A, Shivtiel S, Kalinkovich A et al (2007) Catecholaminergic neurotransmitters regulate migration and repopulation of immature human CD34+ cells through Wnt signaling. Nat Immunol 8:1123–1131. https://doi.org/10.1038/ni1509
Greenbaum A, Hsu YMS, Day RB et al (2013) CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature 495:227–230. https://doi.org/10.1038/nature11926
Friedenstein AJ, Petrakova KV, Kurolesova AI, Frolova GP (1968) Heterotopic of bone marrow. Analysis of precursor cells for osteogenic and hematopoietic tissues. Transplantation 6:230–247
Méndez-Ferrer S, Michurina TV, Ferraro F et al (2010) Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature 466:829–834. https://doi.org/10.1038/nature09262
Asada N, Kunisaki Y, Pierce H et al (2017) Differential cytokine contributions of perivascular haematopoietic stem cell niches. Nat Cell Biol 19:214–223. https://doi.org/10.1038/ncb3475
Sugiyama T, Kohara H, Noda M, Nagasawa T (2006) Maintenance of the hematopoietic stem cell Pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity 25:977–988. https://doi.org/10.1016/j.immuni.2006.10.016
Kunisaki Y, Bruns I, Scheiermann C et al (2013) Arteriolar niches maintain haematopoietic stem cell quiescence. Nature 502:637–643. https://doi.org/10.1038/nature12612
Zhou BO, Yue R, Murphy MM et al (2014) Leptin-receptor-expressing mesenchymal stromal cells represent the main source of bone formed by adult bone marrow. Cell Stem Cell 15:154–168. https://doi.org/10.1016/j.stem.2014.06.008
Bruns I, Lucas D, Pinho S et al (2014) Megakaryocytes regulate hematopoietic stem cell quiescence through CXCL4 secretion. Nat Med 20:1315–1320. https://doi.org/10.1038/nm.3707
Zhao M, Perry JM, Marshall H et al (2014) Megakaryocytes maintain homeostatic quiescence and promote post-injury regeneration of hematopoietic stem cells. Nat Med 20:1321–1326. https://doi.org/10.1038/nm.3706
Pinho S, Marchand T, Yang E et al (2018) Lineage-biased hematopoietic stem cells are regulated by distinct niches. Dev Cell 44:634-641.e4. https://doi.org/10.1016/j.devcel.2018.01.016
Nakamura-Ishizu A, Takubo K, Fujioka M, Suda T (2014) Megakaryocytes are essential for HSC quiescence through the production of thrombopoietin. Biochem Biophys Res Commun 454:353–357. https://doi.org/10.1016/j.bbrc.2014.10.095
Chow A, Lucas D, Hidalgo A et al (2011) Bone marrow CD169+ macrophages promote the retention of hematopoietic stem and progenitor cells in the mesenchymal stem cell niche. J Exp Med 208:761–771. https://doi.org/10.1084/jem.20101688
Ludin A, Itkin T, Gur-Cohen S et al (2012) Monocytes-macrophages that express α-smooth muscle actin preserve primitive hematopoietic cells in the bone marrow. Nat Immunol 13:1072–1082. https://doi.org/10.1038/ni.2408
Hur J, Il CJ, Lee H et al (2016) CD82/KAI1 maintains the dormancy of long-term hematopoietic stem cells through interaction with DARC-expressing macrophages. Cell Stem Cell 18:508–521. https://doi.org/10.1016/j.stem.2016.01.013
Winkler IG, Sims NA, Pettit AR et al (2010) Bone marrow macrophages maintain hematopoietic stem cell (HSC) niches and their depletion mobilizes HSCs. Blood 116:4815–4828. https://doi.org/10.1182/blood-2009-11-253534
Christopher MJ, Rao M, Liu F et al (2011) Expression of the G-CSF receptor in monocytic cells is sufficient to mediate hematopoietic progenitor mobilization by G-CSF in mice. J Exp Med 208:251–260. https://doi.org/10.1084/jem.20101700
Fujisaki J, Wu J, Carlson AL et al (2011) In vivo imaging of T reg cells providing immune privilege to the haematopoietic stem-cell niche. Nature 474:216–220. https://doi.org/10.1038/nature10160
Hirata Y, Furuhashi K, Ishii H et al (2018) CD150 high bone marrow tregs maintain hematopoietic stem cell quiescence and immune privilege via adenosine. Cell Stem Cell 22:445-453.e5. https://doi.org/10.1016/j.stem.2018.01.017
Ding L, Saunders TL, Enikolopov G, Morrison SJ (2012) Endothelial and perivascular cells maintain haematopoietic stem cells. Nature 481:457–462. https://doi.org/10.1038/nature10783
Acar M, Kocherlakota KS, Murphy MM et al (2015) Deep imaging of bone marrow shows non-dividing stem cells are mainly perisinusoidal. Nature 526:126–130. https://doi.org/10.1038/nature15250
Saçma M, Pospiech J, Bogeska R et al (2019) Haematopoietic stem cells in perisinusoidal niches are protected from ageing. Nat Cell Biol 21:1309–1320. https://doi.org/10.1038/s41556-019-0418-y
Xu C, Gao X, Wei Q et al (2018) Stem cell factor is selectively secreted by arterial endothelial cells in bone marrow. Nat Commun 9:1–13. https://doi.org/10.1038/s41467-018-04726-3
Itkin T, Gur-Cohen S, Spencer JA et al (2016) Distinct bone marrow blood vessels differentially regulate haematopoiesis. Nature 532:323–328. https://doi.org/10.1038/nature17624
Kusumbe AP, Ramasamy SK, Itkin T et al (2016) Age-dependent modulation of vascular niches for haematopoietic stem cells. Nature 532:380–384. https://doi.org/10.1038/nature17638
Maryanovich M, Zahalka AH, Pierce H et al (2018) Adrenergic nerve degeneration in bone marrow drives aging of the hematopoietic stem cell niche. Nat Med 24:782–791. https://doi.org/10.1038/s41591-018-0030-x
Ho YH, del Toro R, Rivera-Torres J et al (2019) Remodeling of bone marrow hematopoietic stem cell niches promotes myeloid cell expansion during premature or physiological aging. Cell Stem Cell 25:407-418.e6. https://doi.org/10.1016/j.stem.2019.06.007
Poulos MG, Ramalingam P, Gutkin MC et al (2017) Endothelial transplantation rejuvenates aged hematopoietic stem cell function. J Clin Invest 127:4163–4178. https://doi.org/10.1172/JCI93940
Moerman EJ, Teng K, Lipschitz DA, Lecka-Czernik B (2004) Aging activates adipogenic and suppresses osteogenic programs in mesenchymal marrow stroma/stem cells: the role of PPAR-γ2 transcription factor and TGF-β/BMP signaling pathways. Aging Cell 3:379–389. https://doi.org/10.1111/j.1474-9728.2004.00127.x
Singh L, Brennan TA, Russell E et al (2016) Aging alters bone-fat reciprocity by shifting in vivo mesenchymal precursor cell fate towards an adipogenic lineage. Bone 85:29–36. https://doi.org/10.1016/j.bone.2016.01.014
Nishikawa K, Nakashima T, Takeda S et al (2010) Maf promotes osteoblast differentiation in mice by mediating the age-related switch in mesenchymal cell differentiation. J Clin Invest 120:3455–3465. https://doi.org/10.1172/JCI42528
Fazeli PK, Horowitz MC, MacDougald OA et al (2013) Marrow fat and bone-new perspectives. J Clin Endocrinol Metab 98:935–945. https://doi.org/10.1210/jc.2012-3634
Schwartz AV (2015) Marrow fat and bone: Review of clinical findings. Front Endocrinol (Lausanne) 6:1–6. https://doi.org/10.3389/fendo.2015.00040
Naveiras O, Nardi V, Wenzel PL et al (2009) Bone-marrow adipocytes as negative regulators of the haematopoietic microenvironment. Nature 460:259–263. https://doi.org/10.1038/nature08099
Ambrosi TH, Scialdone A, Graja A et al (2017) Adipocyte accumulation in the bone marrow during obesity and aging impairs stem cell-based hematopoietic and bone regeneration. Cell Stem Cell 20:771-784.e6. https://doi.org/10.1016/j.stem.2017.02.009
Gnani D, Crippa S, della Volpe L et al (2019) An early-senescence state in aged mesenchymal stromal cells contributes to hematopoietic stem and progenitor cell clonogenic impairment through the activation of a pro-inflammatory program. Aging Cell. https://doi.org/10.1111/acel.12933
Liu J, Ding Y, Liu Z, Liang X (2020) Senescence in mesenchymal stem cells: functional alterations, molecular mechanisms, and rejuvenation strategies. Front Cell Dev Biol. https://doi.org/10.3389/fcell.2020.00258
Ho YH, Méndez-Ferrer S (2020) Microenvironmental contributions to hematopoietic stem cell aging. Haematologica 105:38–46. https://doi.org/10.3324/haematol.2018.211334
Ferrucci L, Corsi A, Lauretani F et al (2005) The origins of age-related proinflammatory state. Blood 105:2294–2299. https://doi.org/10.1182/blood-2004-07-2599
Frisch BJ, Hoffman CM, Latchney SE et al (2019) Aged marrow macrophages expand platelet-biased hematopoietic stem cells via interleukin-1B. JCI Insight. https://doi.org/10.1172/jci.insight.124213
Henry CJ, Casás-Selves M, Kim J et al (2015) Aging-associated inflammation promotes selection for adaptive oncogenic events in B cell progenitors. J Clin Invest 125:4666–4680. https://doi.org/10.1172/JCI83024
Valletta S, Thomas A, Meng Y et al (2020) Micro-environmental sensing by bone marrow stroma identifies IL-6 and TGFβ1 as regulators of hematopoietic ageing. Nat Commun. https://doi.org/10.1038/s41467-020-17942-7
Pioli PD, Casero D, Montecino-Rodriguez E et al (2019) Plasma cells are obligate effectors of enhanced myelopoiesis in aging bone marrow. Immunity 51:351-366.e6. https://doi.org/10.1016/j.immuni.2019.06.006
Ergen AV, Boles NC, Goodell MA (2012) Rantes/Ccl5 influences hematopoietic stem cell subtypes and causes myeloid skewing. Blood 119:2500–2509. https://doi.org/10.1182/blood-2011-11-391730
Yamashita M, Passegué E (2019) TNF-α coordinates hematopoietic stem cell survival and myeloid regeneration. Cell Stem Cell 25:357-372.e7. https://doi.org/10.1016/j.stem.2019.05.019
Schürch CM, Riether C, Ochsenbein AF (2014) Cytotoxic CD8+ T cells stimulate hematopoietic progenitors by promoting cytokine release from bone marrow mesenchymal stromal cells. Cell Stem Cell 14:460–472. https://doi.org/10.1016/j.stem.2014.01.002
Pietras EM, Mirantes-Barbeito C, Fong S et al (2016) Chronic interleukin-1 exposure drives haematopoietic stem cells towards precocious myeloid differentiation at the expense of self-renewal. Nat Cell Biol 18:607–618. https://doi.org/10.1038/ncb3346
Essers MAG, Offner S, Blanco-Bose WE et al (2009) IFNα activates dormant haematopoietic stem cells in vivo. Nature 458:904–908. https://doi.org/10.1038/nature07815
Baldridge MT, King KY, Boles NC et al (2010) Quiescent haematopoietic stem cells are activated by IFN-γ in response to chronic infection. Nature 465:793–797. https://doi.org/10.1038/nature09135
Takizawa H, Regoes RR, Boddupalli CS et al (2011) Dynamic variation in cycling of hematopoietic stem cells in steady state and inflammation. J Exp Med 208:273–284. https://doi.org/10.1084/jem.20101643
Esplin BL, Shimazu T, Welner RS et al (2011) Chronic exposure to a TLR ligand injures hematopoietic stem cells. J Immunol 186:5367–5375. https://doi.org/10.4049/jimmunol.1003438
Sato T, Onai N, Yoshihara H et al (2009) Interferon regulatory factor-2 protects quiescent hematopoietic stem cells from type I interferon-dependent exhaustion. Nat Med 15:696–700. https://doi.org/10.1038/nm.1973
Walter D, Lier A, Geiselhart A et al (2015) Exit from dormancy provokes DNA-damage-induced attrition in haematopoietic stem cells. Nature 520:549–552. https://doi.org/10.1038/nature14131
Haas S, Hansson J, Klimmeck D et al (2015) Inflammation-induced emergency megakaryopoiesis driven by hematopoietic stem cell-like megakaryocyte progenitors. Cell Stem Cell 17:422–434. https://doi.org/10.1016/j.stem.2015.07.007
Kaser A, Brandacher G, Steurer W et al (2001) Interleukin-6 stimulates thrombopoiesis through thrombopoietin: role in inflammatory thrombocytosis. Blood 98:2720–2725. https://doi.org/10.1182/blood.V98.9.2720
Matatall KA, Jeong M, Chen S et al (2016) Chronic infection depletes hematopoietic stem cells through stress-induced terminal differentiation. Cell Rep 17:2584–2595. https://doi.org/10.1016/j.celrep.2016.11.031
Zoumbos NC, Gascon P, Djeu JY, Young NS (1985) Interferon is a mediator of hematopoietic suppression in aplastic anemia in vitro and possibly in vivo. Proc Natl Acad Sci USA 82:188–192. https://doi.org/10.1073/pnas.82.1.188
Chen YF, Wu ZM, Xie C et al (2013) Expression level of IL-6 secreted by bone marrow stromal cells in mice with aplastic anemia. ISRN Hematol 2013:1–6. https://doi.org/10.1155/2013/986219
Fuster JJ, MacLauchlan S, Zuriaga MA et al (2017) Clonal hematopoiesis associated with TET2 deficiency accelerates atherosclerosis development in mice. Science 355:842–847. https://doi.org/10.1126/science.aag1381
Ross-Innes CS, Becq J, Warren A et al (2015) Whole-genome sequencing provides new insights into the clonal architecture of Barrett’s esophagus and esophageal adenocarcinoma. Nat Genet 47:1038–1046. https://doi.org/10.1038/ng.3357
Stachler MD, Taylor-Weiner A, Peng S et al (2015) Paired exome analysis of Barrett’s esophagus and adenocarcinoma. Nat Genet 47:1047–1055. https://doi.org/10.1038/ng.3343
Nanki K, Fujii M, Shimokawa M et al (2020) Somatic inflammatory gene mutations in human ulcerative colitis epithelium. Nature 577:254–259. https://doi.org/10.1038/s41586-019-1844-5
Kakiuchi N, Yoshida K, Uchino M et al (2020) Frequent mutations that converge on the NFKBIZ pathway in ulcerative colitis. Nature 577:260–265. https://doi.org/10.1038/s41586-019-1856-1
Brunner SF, Roberts ND, Wylie LA et al (2019) Somatic mutations and clonal dynamics in healthy and cirrhotic human liver. Nature 574:538–542. https://doi.org/10.1038/s41586-019-1670-9
Steensma DP, Bejar R, Jaiswal S et al (2015) Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood 126:9–16. https://doi.org/10.1182/blood-2015-03-631747
Young AL, Challen GA, Birmann BM, Druley TE (2016) Clonal haematopoiesis harbouring AML-associated mutations is ubiquitous in healthy adults. Nat Commun 7:1–7. https://doi.org/10.1038/ncomms12484
McKerrell T, Park N, Moreno T et al (2015) Leukemia-associated somatic mutations drive distinct patterns of age-related clonal hemopoiesis. Cell Rep 10:1239–1245. https://doi.org/10.1016/j.celrep.2015.02.005
Lee-Six H, Øbro NF, Shepherd MS et al (2018) Population dynamics of normal human blood inferred from somatic mutations. Nature 561:473–478. https://doi.org/10.1038/s41586-018-0497-0
Sun J, Ramos A, Chapman B et al (2014) Clonal dynamics of native haematopoiesis. Nature 514:322–327. https://doi.org/10.1038/nature13824
Rodriguez-Fraticelli AE, Wolock SL, Weinreb CS et al (2018) Clonal analysis of lineage fate in native haematopoiesis. Nature 553:212–216. https://doi.org/10.1038/nature25168
Sawai CM, Babovic S, Upadhaya S et al (2016) Hematopoietic stem cells are the major source of multilineage hematopoiesis in adult animals. Immunity 45:597–609. https://doi.org/10.1016/j.immuni.2016.08.007
Pei W, Feyerabend TB, Rössler J et al (2017) Polylox barcoding reveals haematopoietic stem cell fates realized in vivo. Nature 548:456–460. https://doi.org/10.1038/nature23653
Busch K, Klapproth K, Barile M et al (2015) Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature 518:542–546. https://doi.org/10.1038/nature14242
Ganuza M, Hall T, Finkelstein D et al (2019) The global clonal complexity of the murine blood system declines throughout life and after serial transplantation. Blood 133:1927–1942. https://doi.org/10.1182/blood-2018-09-873059
Makishima H, Yoshizato T, Yoshida K et al (2017) Dynamics of clonal evolution in myelodysplastic syndromes. Nat Genet 49:204–212. https://doi.org/10.1038/ng.3742
Walter MJ, Shen D, Ding L et al (2012) Clonal architecture of secondary acute myeloid leukemia. N Engl J Med 366:1090–1098. https://doi.org/10.1056/NEJMoa1106968
Challen GA, Sun D, Mayle A et al (2014) Dnmt3a and Dnmt3b have overlapping and distinct functions in hematopoietic stem cells. Cell Stem Cell 15:350–364. https://doi.org/10.1016/j.stem.2014.06.018
Fujino T, Goyama S, Sugiura Y et al (2021) Mutant ASXL1 induces age-related expansion of phenotypic hematopoietic stem cells through activation of Akt/mTOR pathway. Nat Commun 12:1–20. https://doi.org/10.1038/s41467-021-22053-y
Coombs CC, Zehir A, Devlin SM et al (2017) Therapy-related clonal hematopoiesis in patients with non-hematologic cancers is common and associated with adverse clinical outcomes. Cell Stem Cell 21:374-382.e4. https://doi.org/10.1016/j.stem.2017.07.010
Kahn JD, Miller PG, Silver AJ et al (2018) PPM1D-truncating mutations confer resistance to chemotherapy and sensitivity to PPM1D inhibition in hematopoietic cells. Blood 132:1095–1105. https://doi.org/10.1182/blood-2018-05-850339
Hsu JI, Dayaram T, Tovy A et al (2018) PPM1D mutations drive clonal hematopoiesis in response to cytotoxic chemotherapy. Cell Stem Cell 23:700-713.e6. https://doi.org/10.1016/j.stem.2018.10.004
Bondar T, Medzhitov R (2010) p53-mediated hematopoietic stem and progenitor cell competition. Cell Stem Cell 6:309–322. https://doi.org/10.1016/j.stem.2010.03.002
Hormaechea-Agulla D, Matatall KA, Le DT et al (2021) Chronic infection drives Dnmt3a-loss-of-function clonal hematopoiesis via IFNγ signaling. Cell Stem Cell 28:1428-1442.e6. https://doi.org/10.1016/j.stem.2021.03.002
Higa KC, Goodspeed A, Chavez JS et al (2021) Chronic interleukin-1 exposure triggers selection for Cebpa-knockout multipotent hematopoietic progenitors. J Exp Med. https://doi.org/10.1084/jem.20200560
Meisel M, Hinterleitner R, Pacis A et al (2018) Microbial signals drive pre-leukaemic myeloproliferation in a Tet2-deficient host. Nature 557:580–584. https://doi.org/10.1038/s41586-018-0125-z
Muto T, Walker CS, Choi K et al (2020) Adaptive response to inflammation contributes to sustained myelopoiesis and confers a competitive advantage in myelodysplastic syndrome HSCs. Nat Immunol 21:535–545. https://doi.org/10.1038/s41590-020-0663-z
Zhang Q, Zhao K, Shen Q et al (2015) Tet2 is required to resolve inflammation by recruiting Hdac2 to specifically repress IL-6. Nature 525:389–393. https://doi.org/10.1038/nature15252
Cai Z, Kotzin JJ, Ramdas B et al (2018) Inhibition of inflammatory signaling in Tet2 mutant preleukemic cells mitigates stress-induced abnormalities and clonal hematopoiesis. Cell Stem Cell 23:833-849.e5. https://doi.org/10.1016/j.stem.2018.10.013
Conboy IM, Conboy MJ, Wagers AJ et al (2005) Rejuvenation of aged progenitor cells by exposure to a young systemic environment. Nature 433:760–764. https://doi.org/10.1038/nature03260
Loffredo FS, Steinhauser ML, Jay SM et al (2013) Growth differentiation factor 11 is a circulating factor that reverses age-related cardiac hypertrophy. Cell 153:828–839. https://doi.org/10.1016/j.cell.2013.04.015
Katsimpardi L, Litterman NK, Schein PA et al (2014) Vascular and neurogenic rejuvenation of the aging mouse brain by young systemic factors. Science 344:630–634. https://doi.org/10.1126/science.1251141
Sinha M, Jang YC, Oh J et al (2014) Restoring systemic GDF11 levels reverses age-related dysfunction in mouse skeletal muscle. Science 344:649–652. https://doi.org/10.1126/science.1251152
Ho TT, Dellorusso PV, Verovskaya EV et al (2021) Aged hematopoietic stem cells are refractory to bloodborne systemic rejuvenation interventions. J Exp Med. https://doi.org/10.1084/jem.20210223
Kuribayashi W, Oshima M, Itokawa N et al (2021) Limited rejuvenation of aged hematopoietic stem cells in young bone marrow niche. J Exp Med. https://doi.org/10.1084/jem.20192283
Wahlestedt M, Norddahl GL, Sten G et al (2013) An epigenetic component of hematopoietic stem cell aging amenable to reprogramming into a young state. Blood 121:4257–4264. https://doi.org/10.1182/blood-2012-11-469080
Wahlestedt M, Erlandsson E, Kristiansen T et al (2017) Clonal reversal of ageing-associated stem cell lineage bias via a pluripotent intermediate. Nat Commun. https://doi.org/10.1038/ncomms14533
Nitta E, Itokawa N, Yabata S et al (2020) Bmi1 counteracts hematopoietic stem cell aging by repressing target genes and enforcing the stem cell gene signature. Biochem Biophys Res Commun 521:612–619. https://doi.org/10.1016/j.bbrc.2019.10.153
Agathocleous M, Meacham CE, Burgess RJ et al (2017) Ascorbate regulates haematopoietic stem cell function and leukaemogenesis. Nature 549:476–481. https://doi.org/10.1038/nature23876
Cimmino L, Dolgalev I, Wang Y et al (2017) Restoration of TET2 function blocks aberrant self-renewal and leukemia progression. Cell 170:1079-1095.e20. https://doi.org/10.1016/j.cell.2017.07.032
Cheng CW, Adams GB, Perin L et al (2014) Prolonged fasting reduces IGF-1/PKA to promote hematopoietic-stem-cell- based regeneration and reverse immunosuppression. Cell Stem Cell 14:810–823. https://doi.org/10.1016/j.stem.2014.04.014
Chen C, Liu Y, Liu Y, Zheng P (2009) mTOR regulation and therapeutic rejuvenation of aging hematopoietic stem cells. Sci Signal 2:ra75. https://doi.org/10.1126/scisignal.2000559
Young K, Eudy E, Bell R et al (2021) Decline in IGF1 in the bone marrow microenvironment initiates hematopoietic stem cell aging. Cell Stem Cell 28:1473-1482.e7. https://doi.org/10.1016/j.stem.2021.03.017
Mansell E, Sigurdsson V, Deltcheva E et al (2021) Mitochondrial potentiation ameliorates age-related heterogeneity in hematopoietic stem cell function. Cell Stem Cell 28:241-256.e6. https://doi.org/10.1016/j.stem.2020.09.018
Baker DJ, Wijshake T, Tchkonia T et al (2011) Clearance of p16 Ink4a-positive senescent cells delays ageing-associated disorders. Nature 479:232–236. https://doi.org/10.1038/nature10600
Baar MP, Brandt RMC, Putavet DA et al (2017) Targeted apoptosis of senescent cells restores tissue homeostasis in response to chemotoxicity and aging. Cell 169:132-147.e16. https://doi.org/10.1016/j.cell.2017.02.031
Baker DJ, Childs BG, Durik M et al (2016) Naturally occurring p16 Ink4a-positive cells shorten healthy lifespan. Nature 530:184–189. https://doi.org/10.1038/nature16932
Chang J, Wang Y, Shao L et al (2016) Clearance of senescent cells by ABT263 rejuvenates aged hematopoietic stem cells in mice. Nat Med 22:78–83. https://doi.org/10.1038/nm.4010
Laurenti E, Göttgens B (2018) From haematopoietic stem cells to complex differentiation landscapes. Nature 553:418–426. https://doi.org/10.1038/nature25022
Zhang P, Li X, Pan C et al (2022) Single-cell RNA sequencing to track novel perspectives in HSC heterogeneity. Stem Cell Res Ther 13:1–15. https://doi.org/10.1186/s13287-022-02718-1
Grover A, Sanjuan-Pla A, Thongjuea S et al (2016) Single-cell RNA sequencing reveals molecular and functional platelet bias of aged haematopoietic stem cells. Nat Commun. https://doi.org/10.1038/ncomms11075
Zhong L, Yao L, Tower RJ et al (2020) Single cell transcriptomics identifies a unique adipose lineage cell population that regulates bone marrow environment. Elife 9:1–28. https://doi.org/10.7554/eLife.54695
Almanzar N, Antony J, Baghel AS et al (2020) A single-cell transcriptomic atlas characterizes ageing tissues in the mouse. Nature 583:590–595. https://doi.org/10.1038/s41586-020-2496-1
Miles LA, Bowman RL, Merlinsky TR et al (2020) Single-cell mutation analysis of clonal evolution in myeloid malignancies. Nature 587:477–482. https://doi.org/10.1038/s41586-020-2864-x
Nam AS, Dusaj N, Izzo F et al (2022) Single-cell multi-omics of human clonal hematopoiesis reveals that DNMT3A R882 mutations perturb early progenitor states through selective hypomethylation. BioRxiv. https://doi.org/10.1101/2022.01.14.476225
World health organization (2021) Ageing and health
Pang WW, Price EA, Sahoo D et al (2011) Human bone marrow hematopoietic stem cells are increased in frequency and myeloid-biased with age. Proc Natl Acad Sci USA 108:20012–20017. https://doi.org/10.1073/pnas.1116110108
Funding
This work was supported by Grant-in-Aid for Scientific Research (A) (No. 20H00537) (to T.K.), Grant-in-Aid for Scientific Research on Innovative Areas “Stem Cell Aging and Disease” (No. 17H05634), The Tokyo Biochemical Research Foundation (to T.K.), Japanese Society of Hematology (to T.K.).
Author information
Authors and Affiliations
Contributions
The manuscript was written by TF, SA, SG, and TK took part in writing the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interests
The authors have no relevant financial or non-financial interests to disclose.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent to publish
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Fujino, T., Asada, S., Goyama, S. et al. Mechanisms involved in hematopoietic stem cell aging. Cell. Mol. Life Sci. 79, 473 (2022). https://doi.org/10.1007/s00018-022-04356-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-022-04356-5