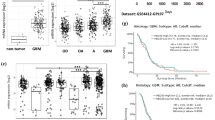
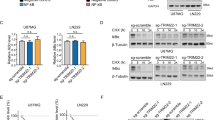
Abstract
Glioblastoma is the most life-threatening tumor of the central nervous system. Despite recent therapeutic advancements, maximum survival of glioblastoma patients remains dismal. The mediator complex is a set of proteins, essential for eukaryotic gene expression. Abnormal expression/mutations of specific mediator genes have been associated with progression of various cancers, however, its role and status in glioblastoma remains largely unknown. Our work shows overexpression of a subunit of kinase assembly of mediator complex, MED12, in various glioblastoma patient cohorts including Indian glioblastoma patients and cell lines. Functional characterization of MED12 using both overexpression and knockdown approach revealed that it promotes glioblastoma cell proliferation, migration and inhibits apoptosis. Transcriptome analysis post MED12 knockdown revealed Vitamin D receptor (VDR) pathway to be one of the key pathways affected by MED12 in glioblastoma. We studied direct interaction of MED12 with VDR protein using docking studies and co-immunoprecipitation assay. We identify BCL6, a secondary regulator of VDR signaling, to be directly regulated by MED12 through a combination of chromatin immunoprecipitation, qRT-PCR and western analyses. We further show that MED12 brings about the inhibition of p53 levels and apoptosis partly through induction of BCL6 in glioblastoma. Overall, this stands as the first report of MED12 over-expression and involvement in glioblastoma pathogenesis and identifies MED12 as an important mediator of VDR signaling and an attractive molecule for development of new therapeutic interventions.








Similar content being viewed by others
Data availability
Upon reasonable request.
Abbreviations
- CDS:
-
Coding sequence
- VDR:
-
Vitamin D receptor
- qRT-PCR:
-
Quantitative real time polymerase chain reaction
- T-ALL:
-
T-cell acute lymphoblastic leukemia
- ESR1:
-
Estrogen receptor alpha
- ESR2:
-
Estrogen receptor beta
- THRA:
-
Thyroid hormone receptor alpha
- siRNA:
-
Small interfering RNA
- MED:
-
Mediator
- MTT:
-
3-(4,5-Dimethylthiazol-2-Yl)-2,5-Diphenyltetrazolium Bromide
- TCGA:
-
The Cancer Genome Atlas
- CGGA:
-
Chinese Glioma Genome Atlas
- GBM:
-
Glioblastoma
- TGFBR2:
-
Transforming growth factor β receptor type 2
References
Donner AJ, Szostek S, Hoover JM, Espinosa JM (2007) CDK8 is a stimulus-specific positive coregulator of p53 target genes. Mol Cell 27(1):121–133. https://doi.org/10.1016/j.molcel.2007.05.026
Galbraith MD, Donner AJ, Espinosa JM (2010) CDK8: a positive regulator of transcription. Transcription 1(1):4–12. https://doi.org/10.4161/trns.1.1.12373
Fj A, Yw J, Lc M, Cm G, Rd K (1999) Conserved structures of mediator and RNA polymerase II holoenzyme. Science (New York, NY) 283:5404. https://doi.org/10.1126/science.283.5404.985
Daniels L, D. (2013) Mutual exclusivity of MED12/MED12L, MED13/13L, and CDK8/19 paralogs revealed within the CDK-mediator kinase module. J Proteom Bioinform 01:S2. https://doi.org/10.4172/jpb.S2-004
Allen BL, Taatjes DJ (2015) The mediator complex: a central integrator of transcription. Nat Rev Mol Cell Biol 16(3):155. https://doi.org/10.1038/nrm3951
Fant CB, Taatjes DJ (2018) Regulatory functions of the mediator kinases CDK8 and CDK19. Transcription 10(2):76–90. https://doi.org/10.1080/21541264.2018.1556915
Hirst M, Kobor MS, Kuriakose N, Greenblatt J, Sadowski I (1999) GAL4 is regulated by the RNA polymerase II holoenzyme-associated cyclin-dependent protein kinase SRB10/CDK8. Mol Cell 3(5):673–678. https://doi.org/10.1016/S1097-2765(00)80360-3
Zhao X, Feng D, Wang Q, Abdulla A, Xie X-J, Zhou J, Sun Y, Yang ES, Liu L-P, Vaitheesvaran B, Bridges L, Kurland IJ, Strich R, Ni J-Q, Wang C, Ericsson J, Pessin JE, Ji J-Y, Yang F (2012) Regulation of lipogenesis by cyclin-dependent kinase 8-mediated control of SREBP-1. J Clin Investig 122(7):2417. https://doi.org/10.1172/JCI61462
Cj H, Ve M, Sm L, Cj W, Ss K, Ra Y (1998) Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol Cell 2:1. https://doi.org/10.1016/s1097-2765(00)80112-4
Akoulitchev S, Chuikov S, Reinberg D (2000) TFIIH is negatively regulated by cdk8-containing mediator complexes. Nature 407(6800):102–106. https://doi.org/10.1038/35024111
Liang J, Chen M, Hughes D, Chumanevich AA, Altilia S, Kaza V, Lim C-U, Kiaris H, Mythreye K, Pena MM, Broude EV, Roninson IB (2018) CDK8 selectively promotes the growth of colon cancer metastases in the liver by regulating gene expression of TIMP3 and matrix metalloproteinases. Can Res 78(23):6594–6606. https://doi.org/10.1158/0008-5472.CAN-18-1583
Bancerek J, Poss ZC, Steinparzer I, Sedlyarov V, Pfaffenwimmer T, Mikulic I, Dölken L, Strobl B, Müller M, Taatjes DJ, Kovarik P (2013) CDK8 kinase phosphorylates transcription factor STAT1 to selectively regulate the interferon response. Immunity 38(2):250. https://doi.org/10.1016/j.immuni.2012.10.017
Alarcón C, Zaromytidou A-I, Xi Q, Gao S, Yu J, Fujisawa S, Barlas A, Miller AN, Manova-Todorova K, Macias MJ, Sapkota G, Pan D, Massagué J (2009) Nuclear CDKs drive smad transcriptional activation and turnover in BMP and TGF-β pathways. Cell 139(4):757–769. https://doi.org/10.1016/j.cell.2009.09.035
Poss ZC, Ebmeier CC, Taatjes DJ (2013) The mediator complex and transcription regulation. Crit Rev Biochem Mol Biol. https://doi.org/10.3109/10409238.2013.840259
Srivastava S, Kulshreshtha R (2020) Insights into the regulatory role and clinical relevance of mediator subunit, MED12, in human diseases. J Cell Physiol 236(5):3163–3177. https://doi.org/10.1002/jcp.30099
Zhang S, O’Regan R, Xu W (2020) The emerging role of mediator complex subunit 12 in tumorigenesis and response to chemotherapeutics. Cancer 126(5):939–948. https://doi.org/10.1002/cncr.32672
El Andaloussi A, Al-Hendy A, Ismail N, Boyer TG, Halder SK (2020) Introduction of somatic mutation in MED12 induces Wnt4/β-catenin and disrupts autophagy in human uterine myometrial cell. Reprod Sci 27:823–832. https://doi.org/10.1007/s43032-019-00084-7
Wu B, Słabicki M, Sellner L, Dietrich S, Liu X, Jethwa A, Hüllein J, Walther T, Wagner L, Huang Z, Zapatka M, Zenz T (2017) MED12 mutations and NOTCH signalling in chronic lymphocytic leukaemia. Br J Haematol 179:421–429. https://doi.org/10.1111/bjh.14869
Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat J-P, White TA, Stojanov P, Van Allen E, Stransky N, Nickerson E, Chae S-S, Boysen G, Auclair D, Onofrio RC, Park K, Kitabayashi N, MacDonald TY, Sheikh K, Garraway LA (2012) Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet 44:685–689. https://doi.org/10.1038/ng.2279
Luo X-L, Deng C, Su X, Wang F, Chen Z, Wu X-P, Liang S-B, Liu J, Fu L (2018) Loss of MED12 induces tumour dormancy in human epithelial ovarian cancer via downregulation of EGFR. Cancer Res. https://doi.org/10.1158/0008-5472.CAN-18-0134
Wang L, Zeng H, Wang Q, Zhao Z, Boyer TG, Bian X, Xu W (2015) MED12 methylation by CARM1 sensitizes human breast cancer cells to chemotherapy drugs. Sci Adv 1:e1500463. https://doi.org/10.1126/sciadv.1500463
Ciebiera M, Ali M, Zgliczyńska M, Skrzypczak M, Al-Hendy A (2020) Vitamins and uterine fibroids: current data on pathophysiology and possible clinical relevance. Int J Mol Sci 21(15):5528. https://doi.org/10.3390/ijms21155528
Xu M, Wang F, Li G, Wang X, Fang X, Jin H, Chen Z, Zhang J, Fu L (2019) MED12 exerts an emerging role in actin-mediated cytokinesis via LIMK2/cofilin pathway in NSCLC. Mol Cancer 18:93. https://doi.org/10.1186/s12943-019-1020-4
Huang S, Hölzel M, Knijnenburg T, Schlicker A, Roepman P, McDermott U, Garnett M, Grernrum W, Sun C, Prahallad A, Groenendijk FH, Mittempergher L, Nijkamp W, Neefjes J, Salazar R, ten Dijke P, Uramoto H, Tanaka F, Beijersbergen RL, Wessels LFA, Bernards R (2012) MED12 controls the response to multiple cancer drugs through regulation of TGF-β receptor signalling. Cell 151:937–950. https://doi.org/10.1016/j.cell.2012.10.035
Shaikhibrahim Z, Offermann A, Braun M, Menon R, Syring I, Nowak M, Halbach R, Vogel W, Ruiz C, Zellweger T, Rentsch CA, Svensson M, Andren O, Bubendorf L, Biskup S, Duensing S, Kirfel J, Perner S (2014) MED12 overexpression is a frequent event in castration-resistant prostate cancer. Endocr Relat Cancer 21:663–675. https://doi.org/10.1530/ERC-14-0171
Crown J (2017) CDK8: A new breast cancer target. Oncotarget 8(9):14269. https://doi.org/10.18632/oncotarget.15354
Gu W, Wang C, Li W, Hsu F-N, Tian L, Zhou J, Yuan C, Xie X-J, Jiang T, Addya S, Tai Y, Kong B, Ji J-Y (2013) Tumor-suppressive effects of CDK8 in endometrial cancer cells. Cell Cycle 12(6):987–999. https://doi.org/10.4161/cc.24003
Kapoor A, Goldberg MS, Cumberland LK, Ratnakumar K, Segura MF, Emanuel PO, Menendez S, Vardabasso C, LeRoy G, Vidal CI, Polsky D, Osman I, Garcia BA, Hernando E, Bernstein E (2010) The histone variant macroH2A suppresses melanoma progression through regulation of CDK8. Nature 468(7327):1105. https://doi.org/10.1038/nature09590
Li N, Fassl A, Chick J, Inuzuka H, Li X, Mansour MR, Liu L, Wang H, King B, Shaik S, Gutierrez A, Ordureau A, Otto T, Kreslavsky T, Baitsch L, Bury L, Meyer CA, Ke N, Mulry KA, Sicinski P (2014) Cyclin C is a haploinsufficient tumour suppressor. Nat Cell Biol 16(11):1080–1091. https://doi.org/10.1038/ncb3046
Ohata N, Ito S, Yoshida A, Kunisada T, Numoto K, Jitsumori Y, Kanzaki H, Ozaki T, Shimizu K, Ouchida M (2006) Highly frequent allelic loss of chromosome 6q16–23 in osteosarcoma: Involvement of cyclin C in osteosarcoma. Int J Mol Med 18(6). https://pubmed.ncbi.nlm.nih.gov/17089020/
Thakkar JP, Dolecek TA, Horbinski C, Ostrom QT, Lightner DD, Barnholtz-Sloan JS, Villano JL (2014) Epidemiologic and molecular prognostic review of glioblastoma. Cancer Epidemiol Biomark Prev Publ Am Assoc Cancer Res 23:10. https://doi.org/10.1158/1055-9965.EPI-14-0275
Stupp R, Mason WP, van Bent MJ, Weller M, Fisher B, Taphoorn MJB, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO (2009) Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. Mass Med Soc. https://doi.org/10.1056/NEJMoa043330
Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, Parada LF (2012) A restricted cell population propagates glioblastoma growth after chemotherapy. Nature 488:7412. https://doi.org/10.1038/nature11287
Lee G, Auffinger B, Guo D, Hasan T, Deheeger M, Tobias AL, Kim JY, Atashi F, Zhang L, Lesniak MS, James CD (2016) Dedifferentiation of glioma cells to glioma stem-like cells by therapeutic stress-induced HIF signaling in the recurrent GBM model. Mol Cancer Therap 15:12. https://doi.org/10.1158/1535-7163.MCT-15-0675
Noch EK, Ramakrishna R, Magge R (2018) Challenges in the treatment of glioblastoma: multisystem mechanisms of therapeutic resistance. World Neurosurg 116:505–517. https://doi.org/10.1016/j.wneu.2018.04.022
Shukla A, Srivastava S, Darokar J, Kulshreshtha R (2020) HIF1α and p53 regulated MED30, a mediator complex subunit, is involved in regulation of glioblastoma pathogenesis and temozolomide resistance. Cell Mol Neurobiol. https://doi.org/10.1007/s10571-020-00920-4
Xu L, Chen Y, Dutra-Clarke M, Mayakonda A, Hazawa M, Savinoff SE, Doan N, Said JW, Yong WH, Watkins A, Yang H, Ding L-W, Jiang Y-Y, Tyner JW, Ching J, Kovalik J-P, Madan V, Chan S-L, Müschen M, Koeffler HP (2017) BCL6 promotes glioma and serves as a therapeutic target. Proc Natl Acad Sci USA 114(15):3981. https://doi.org/10.1073/pnas.1609758114
Fabre M-S, Stanton NM, Slatter TL, Lee S, Senanayake D, Gordon RMA, Castro ML, Rowe MR, Taha A, Royds JA, Hung N, Melnick AM, McConnell MJ (2020) The oncogene BCL6 is up-regulated in glioblastoma in response to DNA damage, and drives survival after therapy. PLoS One 15(4):e0231470. https://doi.org/10.1371/journal.pone.0231470
Clark AD, Oldenbroek M, Boyer TG (2015) Mediator kinase module and human tumorigenesis. Crit Rev Biochem Mol Biol 50(5):393. https://doi.org/10.3109/10409238.2015.1064854
Chen J-R, Yao Y, Xu H-Z, Qin Z-Y (2016) Isocitrate dehydrogenase (IDH)1/2 mutations as prognostic markers in patients with glioblastomas. Medicine 95(9):e2583. https://doi.org/10.1097/MD.0000000000002583
Chen W, Roeder RG (2011) Mediator-dependent nuclear receptor function. Semin Cell Dev Biol 22(7):749–758. https://doi.org/10.1016/j.semcdb.2011.07.026
Nagpal N, Sharma S, Maji S, Durante G, Ferracin M, Thakur JK, Kulshreshtha R (2018) Essential role of MED1 in the transcriptional regulation of ER-dependent oncogenic miRNAs in breast cancer. Sci Rep 8(1):11805. https://doi.org/10.1038/s41598-018-29546-9
Nurminen V, Neme A, Ryynanen J, Heikkinen S, Seuter S, Carlberg C (2015) The transcriptional regulator BCL6 participates in the secondary gene regulatory response to vitamin D. Biochim Biophys Acta 1849:3. https://doi.org/10.1016/j.bbagrm.2014.12.001
Vukić M, Neme A, Seuter S, Saksa N, de Mello VDF, Nurmi T, Uusitupa M, Tuomainen T-P, Virtanen JK, Carlberg C (2015) Relevance of vitamin D receptor target genes for monitoring the vitamin D responsiveness of primary human cells. PLoS One 10(4):e0124339. https://doi.org/10.1371/journal.pone.0124339
Warwick T, Schulz MH, Günther S, Gilsbach R, Neme A, Carlberg C, Brandes RP, Seuter S (2021) A hierarchical regulatory network analysis of the vitamin D induced transcriptome reveals novel regulators and complete VDR dependency in monocytes. Sci Rep 11(1):1–16. https://doi.org/10.1038/s41598-021-86032-5
Zhang Y, Dube C, Gibert M, Cruickshanks N, Wang B, Coughlan M, Yang Y, Setiady I, Deveau C, Saoud K, Grello C (2018) The p53 pathway in glioblastoma. Cancers 10:9. https://doi.org/10.3390/cancers10090297
Bowman RL, Wang Q, Carro A, Verhaak RGW, Squatrito M (2017) GlioVis data portal for visualization and analysis of brain tumor expression datasets. Neuro Oncol 19(1):139. https://doi.org/10.1093/neuonc/now247
Mizuno H, Kitada K, Nakai K, Sarai A (2009) PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC Med Genom 2(1):1–11. https://doi.org/10.1186/1755-8794-2-18
Thul PJ, Lindskog C (2018) The human protein atlas: a spatial map of the human proteome. Prot Sci Publ Prot Soc 27(1):233. https://doi.org/10.1002/pro.3307
Stark C, Breitkreutz B-J, Reguly T, Boucher L, Breitkreutz A, Tyers M (2006) BioGRID: a general repository for interaction datasets. Nucleic Acids Res 34(Database issue):D535. https://doi.org/10.1093/nar/gkj109
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6:l1. https://doi.org/10.1126/scisignal.2004088
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR (2010) A method and server for predicting damaging missense mutations. Nat Methods 7(4):248. https://doi.org/10.1038/nmeth0410-248
Acknowledgements
SS thanks Ministry of Human Resource and Development (MHRD), Govt. of India for senior research fellowship. HM thanks Department of Science and Technology, Govt. of India for post-doctoral fellowship. VS thanks Department of Biotechnology, Govt. of India for the post-doctoral fellowship. CS thanks Science and Engineering Research Board (SERB), Govt. of India for the JC Bose fellowship.
Funding
RK and CS thank Department of Biotechnology (DBT), Government of India for financial support (BT/PR16851/MED/122/45/2016). CS also thanks the Science and Engineering Research Board, Govt. of India for the JC Bose fellowship.
Author information
Authors and Affiliations
Contributions
RK conceptualized and coordinated the whole study. SS performed all the cell line experiments and data analyses of patient data. HM and SS performed co-immunoprecipitation experiments. HM performed docking studies.VS1, VS2 and CS performed analyses in the Indian GBM Patient samples. SS, HM and RK wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing financial or other interest in relation to this work.
Ethics approval
The study was approved by the Ethics committee (Ref. No. IEC-130/07.04.2017, RP-24/2017) of All India Institute of Medical Sciences, New Delhi, and informed consent was obtained from the patients.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
18_2021_4056_MOESM2_ESM.xlsx
Supplementary file2 List of differentially regulated genes (p-value < 0.05, fold change > or = + -1.5) obtained by gene expression profiling in A172 cells upon MED12 knockdown (XLSX 31 KB)
18_2021_4056_MOESM3_ESM.xlsx
Supplementary file3 Results of pathway analysis of differentially regulated genes (p-value < 0.05, fold change > or = + -1.5) obtained by gene expression profiling in A172 cells upon MED12 knockdown (XLSX 15 KB)
18_2021_4056_MOESM4_ESM.docx
Supplementary file4 Supplementary tables with list of primers, list of amino acids in MED12 with VDR having hydrogen bond interactions and list of amino acids in MED12 with VDR having hydrophobic interactions. (DOCX 18 KB)
18_2021_4056_MOESM5_ESM.tif
Supplementary file5 Supplementary Fig. 1: (1–10) IHC Images of glioma patients (antibody used: HPA003184) showing medium (n = 2) to high (n = 8) expression in MED12 protein. Images were retrieved from the Human Protein Atlas (TIF 21424 KB)
18_2021_4056_MOESM6_ESM.tif
Supplementary file6 Supplementary Fig. 2: MED12 transcript levels were measured 48 h post-transfection in GBM cells (a) qRT-PCR data showing knockdown of MED12 transcript levels in A172 cell line upon transfection with MED12 specific siRNA (Sigma). (b) qRT-PCR data show induction in MED12 transcript levels upon transfection with MED12 over-expression construct in A172 cell line. (c) qRT-PCR data showing knockdown of MED12 transcript levels in T98G cell line upon transfection with MED12 specific siRNA (Sigma). (d) qRT-PCR data shows induction in MED12 transcript levels upon transfection with MED12 over-expression construct in T98G cell line (TIF 21424 KB)
18_2021_4056_MOESM7_ESM.tif
Supplementary file7 Supplementary Fig. 3: (a, b) Graph showing fold change in expression of genes of VDR pathway analysed by qRT-PCR post MED12 over-expression in (a) A172 and (b) T98G cells (TIF 21424 KB)
18_2021_4056_MOESM8_ESM.tif
Supplementary file8 Supplementary Fig. 4: MED12 is enhanced in IDH mutant tumors. Data analysis from GlioVis showed enhanced expression of MED12 in IDH mutant tumors as compared to IDH wild type tumors across various datasets (a) analysis of CGGA patient dataset (b) analysis of TCGA_GBMLGG patient dataset (c) analysis of Gravendeel patient dataset (d) analysis of Bao patient dataset (TIF 21424 KB)
18_2021_4056_MOESM9_ESM.tif
Supplementary file9 Supplementary Fig. 5: BCL6 inhibits p53 expression in GBM cells. Western Blotting data to show inhibition in p53 levels post BCL6 overexpression in (a) A172 cells (b) T98G cells. The western blotting experiment was performed in duplicates. The actin blot was run on a different gel (TIF 7142 KB)
18_2021_4056_MOESM10_ESM.tif
Supplementary file10 Supplementary Fig. 6: Inhibition of apoptosis by MED12 is BCL6 mediated: Simultaneous over-expression of BCL6 and MED12 knockdown was performed in A172 and T98G cells. (a, b) Western blotting results showing effects of MED12 knockdown and BCL6 over-expression alone or in combination on cleaved PARP levels in (a) A172 cells (b) T98G cells. The western blotting experiment was performed in duplicates. (c) FACS based detection of phosphatidylserine externalisation was checked in A172 cells post simultaneous over-expression of BCL6 and MED12 (TIF 21424 KB)
Rights and permissions
About this article
Cite this article
Srivastava, S., Makala, H., Sharma, V. et al. MED12 is overexpressed in glioblastoma patients and serves as an oncogene by targeting the VDR/BCL6/p53 axis. Cell. Mol. Life Sci. 79, 104 (2022). https://doi.org/10.1007/s00018-021-04056-6
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-021-04056-6