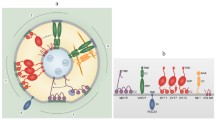
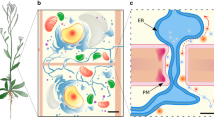
Abstract
Plasma membranes are heterogeneous and laterally compartmentalized into distinct microdomains. These membrane microdomains consist of special lipids and proteins and are thought to act as signaling platforms. In plants, membrane microdomains have been detected by super-resolution microscopy, and there is evidence that they play roles in several biological processes. Here, we review current knowledge about the lipid and protein components of membrane microdomains. Furthermore, we summarize the dynamics of membrane microdomains in response to different stimuli. We also explore the biological functions associated with membrane microdomains as signal integration hubs. Finally, we outline challenges and questions for further studies.


Similar content being viewed by others
References
Lichtenberg D, Goni FM, Heerklotz H (2005) Detergent-resistant membranes should not be identified with membrane rafts. Trends Biochem Sci 30:430–436
Singer SJ, Nicolson GL (1972) The fluid mosaic model of the structure of cell membranes. Science 175:720–731
Yu J, Fischman DA, Steck TL (1973) Selective solubilization of proteins and phospholipids from red blood cell membranes by nonionic detergents. J Supramol Struct 1:233–248
Varma R, Mayor S (1998) GPI-anchored proteins are organized in submicron domains at the cell surface. Nature 394:798–801
Pralle A, Keller P, Florin EL, Simons K, Horber JK (2000) Sphingolipid-cholesterol rafts diffuse as small entities in the plasma membrane of mammalian cells. J Cell Biol 148:997–1008
Friedrichson T, Kurzchalia TV (1998) Microdomains of GPI-anchored proteins in living cells revealed by crosslinking. Nature 394:802–805
Simons K, van Meer G (1988) Lipid sorting in epithelial cells. Biochemistry 27:6197–6202
Simons K, Vaz WLC (2004) Model systems, lipid rafts, and cell membranes. Annu Rev Biophys Biomol Struct 33:269–295
Jacobson K, Mouritsen OG, Anderson RGW (2007) Lipid rafts: at a crossroad between cell biology and physics. Nat Cell Biol 9:7–14
Malinsky J, Opekarova M, Tanner W (2010) The lateral compartmentation of the yeast plasma membrane. Yeast 27:473–478
Mongrand S, Stanislas T, Bayer EMF, Lherminier J, Simon-Plas F (2010) Membrane rafts in plant cells. Trends Plant Sci 15:656–663
Simons K, Gerl MJ (2010) Revitalizing membrane rafts: new tools and insights. Nat Rev Mol Cell Bio 11:688–699
Sezgin E, Levental I, Mayor S, Eggeling C (2017) The mystery of membrane organization: composition, regulation and roles of lipid rafts. Nat Rev Mol Cell Bio 18:361–374
Brown DA, Rose JK (1992) Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell 68:533–544
Shogomori H, Brown DA (2003) Use of detergents to study membrane rafts: the good, the bad, and the ugly. Biol Chem 384:1259–1263
Malinsky J, Opekarova M, Grossmann G, Tanner W (2013) Membrane microdomains, rafts, and detergent-resistant membranes in plants and fungi. Annu Rev Plant Bio 64:501–529
Sezgin E, Schwille P (2012) Model membrane platforms to study protein-membrane interactions. Mol Membr Biol 29:144–154
Day CA, Kenworthy AK (2009) Tracking microdomain dynamics in cell membranes. Biochim Biophys Acta 1788:245–253
Lagerholm BC, Weinreb GE, Jacobson K, Thompson NL (2005) Detecting microdomains in intact cell membranes. Annu Rev Phys Chem 56:309–336
Yang K, Cheng H, Gross RW, Han X (2009) Automated lipid identification and quantification by multidimensional mass spectrometry-based shotgun lipidomics. Anal Chem 81:4356–4368
Herzog R, Schwudke D, Schuhmann K, Sampaio JL, Bornstein SR, Schroeder M, Shevchenko A (2011) A novel informatics concept for high-throughput shotgun lipidomics based on the molecular fragmentation query language. Genome Biol 12:R8
Haimi P, Uphoff A, Hermansson M, Somerharju P (2006) Software tools for analysis of mass spectrometric lipidome data. Anal Chem 78:8324–8331
Mongrand S, Morel J, Laroche J, Claverol S, Carde JP, Hartmann MA, Bonneu M, Simon-Plas F, Lessire R, Bessoule JJ (2004) Lipid rafts in higher plant cells: purification and characterization of Triton X-100-insoluble microdomains from tobacco plasma membrane. J Biol Chem 279:36277–36286
Laloi M, Perret AM, Chatre L, Melser S, Cantrel C, Vaultier MN, Zachowski A, Bathany K, Schmitter JM, Vallet M et al (2007) Insights into the role of specific lipids in the formation and delivery of lipid microdomains to the plasma membrane of plant cells. Plant Physiol 143:461–472
Cacas JL, Furt F, Le Guedard M, Schmitter JM, Bure C, Gerbeau-Pissot P, Moreau P, Bessoule JJ, Simon-Plas F, Mongrand S (2012) Lipids of plant membrane rafts. Prog Lipid Res 51:272–299
Guo DA, Venkatramesh M, Nes WD (1995) Developmental regulation of sterol biosynthesis in Zea mays. Lipids 30:203–219
Travis RL, Berkowitz RL (1980) Characterization of soybean plasma membrane during development: free sterol composition and concanavalin a binding studies. Plant Physiol 65:871–879
Norberg P, Liljenberg C (1991) Lipids of plasma membranes prepared from oat root cells: effects of induced water-deficit tolerance. Plant Physiol 96:1136–1141
Grandmougin A, Bouvier-Nave P, Ullmann P, Benveniste P, Hartmann MA (1989) Cyclopropyl sterol and phospholipid composition of membrane fractions from maize roots treated with fenpropimorph. Plant Physiol 90:591–597
Pata MO, Hannun YA, Ng CKY (2010) Plant sphingolipids: decoding the enigma of the Sphinx. New Phytol 185:611–630
Cacas JL, Bure C, Furt F, Maalouf JP, Badoc A, Cluzet S, Schmitter JM, Antajan E, Mongrand S (2013) Biochemical survey of the polar head of plant glycosylinositolphosphoceramides unravels broad diversity. Phytochemistry 96:191–200
Sperling P, Heinz E (2003) Plant sphingolipids: structural diversity, biosynthesis, first genes and functions. Bba-Mol Cell Biol L 1632:1–15
Cacas JL, Bure C, Grosjean K, Gerbeau-Pissot P, Lherminier J, Rombouts Y, Maes E, Bossard C, Gronnier J, Furt F et al (2016) Revisiting plant plasma membrane lipids in tobacco: a focus on sphingolipids. Plant Physiol 170:367–384
Furt F, Konig S, Bessoule JJ, Sargueil F, Zallot R, Stanislas T, Noirot E, Lherminier J, Simon-Plas F, Heilmann I et al (2010) Polyphosphoinositides are enriched in plant membrane rafts and form microdomains in the plasma membrane. Plant Physiol 152:2173–2187
van Meer G, Stelzer EH, Wijnaendts-van-Resandt RW, Simons K (1987) Sorting of sphingolipids in epithelial (Madin-Darby canine kidney) cells. J Cell Biol 105:1623–1635
Lentz BR, Burgess SW (1989) A dimerization model for the concentration dependent photophysical properties of diphenylhexatriene and its phospholipid derivatives. DPHpPC and DPHpPA. Biophys J 56:723–733
Kusube M, Tamai N, Matsuki H, Kaneshina S (2005) Pressure-induced phase transitions of lipid bilayers observed by fluorescent probes Prodan and Laurdan. Biophys Chem 117:199–206
Jin L, Millard AC, Wuskell JP, Dong X, Wu D, Clark HA, Loew LM (2006) Characterization and application of a new optical probe for membrane lipid domains. Biophys J 90:2563–2575
Beknke O, Tranum-Jensen J, van Deurs B (1984) Filipin as a cholesterol probe. I. Morphology of filipin-cholesterol interaction in lipid model systems. Eur J Cell Biol 35:189–199
Singh RD, Liu Y, Wheatley CL, Holicky EL, Makino A, Marks DL, Kobayashi T, Subramaniam G, Bittman R, Pagano RE (2006) Caveolar endocytosis and microdomain association of a glycosphingolipid analog is dependent on its sphingosine stereochemistry. J Biol Chem 281:30660–30668
Le Guyader L, Le Roux C, Mazeres S, Gaspard-Iloughmane H, Gornitzka H, Millot C, Mingotaud C, Lopez A (2007) Changes of the membrane lipid organization characterized by means of a new cholesterol-pyrene probe. Biophys J 93:4462–4473
Konigshofer H, Tromballa HW, Loppert HG (2008) Early events in signalling high-temperature stress in tobacco BY2 cells involve alterations in membrane fluidity and enhanced hydrogen peroxide production. Plant Cell Environ 31:1771–1780
Liu P, Li RL, Zhang L, Wang QL, Niehaus K, Baluska F, Samaj J, Lin JX (2009) Lipid microdomain polarization is required for NADPH oxidase-dependent ROS signaling in Picea meyeri pollen tube tip growth. Plant J 60:303–313
Zhao X, Li R, Lu C, Baluska F, Wan Y (2015) Di-4-ANEPPDHQ, a fluorescent probe for the visualisation of membrane microdomains in living Arabidopsis thaliana cells. Plant Physiol Biochem 87:53–60
Li R, Liu P, Wan Y, Chen T, Wang Q, Mettbach U, Baluska F, Samaj J, Fang X, Lucas WJ et al (2012) A membrane microdomain-associated protein, Arabidopsis Flot1, is involved in a clathrin-independent endocytic pathway and is required for seedling development. Plant Cell 24:2105–2122
Stauffer TP, Ahn S, Meyer T (1998) Receptor-induced transient reduction in plasma membrane PtdIns(4,5)P2 concentration monitored in living cells. Curr Biol 8:343–346
Varnai P, Balla T (1998) Visualization of phosphoinositides that bind pleckstrin homology domains: calcium- and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. J Cell Biol 143:501–510
van Leeuwen W, Vermeer JE, Gadella Jr TW, Munnik T. (2007) Visualization of phosphatidylinositol 4,5-bisphosphate in the plasma membrane of suspension-cultured tobacco BY-2 cells and whole Arabidopsis seedlings. Plant J 52:1014–1026
Demir F, Horntrich C, Blachutzik JO, Scherzer S, Reinders Y, Kierszniowska S, Schulze WX, Harms GS, Hedrich R, Geiger D et al (2013) Arabidopsis nanodomain-delimited ABA signaling pathway regulates the anion channel SLAH3. Proc Natl Acad Sci USA 110:8296–8301
Hao H, Fan L, Chen T, Li R, Li X, He Q, Botella MA, Lin J (2014) Clathrin and membrane microdomains cooperatively regulate rbohd dynamics and activity in Arabidopsis. Plant Cell 26:1729–1745
Li X, Wang X, Yang Y, Li R, He Q, Fang X, Luu DT, Maurel C, Lin J (2011) Single-molecule analysis of PIP2;1 dynamics and partitioning reveals multiple modes of Arabidopsis plasma membrane aquaporin regulation. Plant Cell 23:3780–3797
Keinath NF, Kierszniowska S, Lorek J, Bourdais G, Kessler SA, Shimosato-Asano H, Grossniklaus U, Schulze WX, Robatzek S, Panstruga R (2010) PAMP (pathogen-associated molecular pattern)-induced changes in plasma membrane compartmentalization reveal novel components of plant immunity. J Biol Chem 285:39140–39149
Takahashi A, Kawasaki T, Wong HL, Suharsono U, Hirano H, Shimamoto K (2003) Hyperphosphorylation of a mitochondrial protein, prohibitin, is induced by calyculin A in a rice lesion-mimic mutant cdr1. Plant Physiol 132:1861–1869
Ahn CS, Lee JH, Reum Hwang A, Kim WT, Pai HS (2006) Prohibitin is involved in mitochondrial biogenesis in plants. Plant J 46:658–667
Borner GH, Sherrier DJ, Weimar T, Michaelson LV, Hawkins ND, Macaskill A, Napier JA, Beale MH, Lilley KS, Dupree P (2005) Analysis of detergent-resistant membranes in Arabidopsis. Evidence for plasma membrane lipid rafts. Plant Physiol 137:104–116
Winzer T, Bairl A, Linder M, Linder D, Werner D, Muller P (1999) A novel 53-kDa nodulin of the symbiosome membrane of soybean nodules, controlled by Bradyrhizobium japonicum. Mol Plant Microbe Interact 12:218–226
Saalbach G, Erik P, Wienkoop S (2002) Characterisation by proteomics of peribacteroid space and peribacteroid membrane preparations from pea (Pisum sativum) symbiosomes. Proteomics 2:325–337
Nadimpalli R, Yalpani N, Johal GS, Simmons CR (2000) Prohibitins, stomatins, and plant disease response genes compose a protein superfamily that controls cell proliferation, ion channel regulation, and death. J Biol Chem 275:29579–29586
Rostoks N, Schmierer D, Kudrna D, Kleinhofs A (2003) Barley putative hypersensitive induced reaction genes: genetic mapping, sequence analyses and differential expression in disease lesion mimic mutants. Theor Appl Genet 107:1094–1101
Raffaele S, Mongrand S, Gamas P, Niebel A, Ott T (2007) Genome-wide annotation of remorins, a plant-specific protein family: evolutionary and functional perspectives. Plant Physiol 145:593–600
Konrad SS, Ott T (2015) Molecular principles of membrane microdomain targeting in plants. Trends Plant Sci 20:351–361
Wang L, Xue Y, Xing J, Song K, Lin J (2018) Exploring the spatiotemporal organization of membrane proteins in living plant cells. Annu Rev Plant Biol 69:525–551
Yu M, Liu H, Dong Z, Xiao J, Su B, Fan L, Komis G, Samaj J, Lin J, Li R (2017) The dynamics and endocytosis of Flot1 protein in response to flg22 in Arabidopsis. J Plant Physiol 215:73–84
Jarsch IK, Konrad SS, Stratil TF, Urbanus SL, Szymanski W, Braun P, Braun KH, Ott T (2014) Plasma membranes are subcompartmentalized into a plethora of coexisting and diverse microdomains in Arabidopsis and Nicotiana benthamiana. Plant Cell 26:1698–1711
Hodzic A, Rappolt M, Amenitsch H, Laggner P, Pabst G (2008) Differential modulation of membrane structure and fluctuations by plant sterols and cholesterol. Biophys J 94:3935–3944
Garcia-Saez AJ, Schwille P (2010) Stability of lipid domains. FEBS Lett 584:1653–1658
Gui J, Zheng S, Liu C, Shen J, Li J, Li L (2016) OsREM4.1 Interacts with OsSERK1 to coordinate the Interlinking between abscisic acid and brassinosteroid signaling in rice. Dev Cell 38:201–213
Konrad SS, Popp C, Stratil TF, Jarsch IK, Thallmair V, Folgmann J, Marin M, Ott T (2014) S-acylation anchors remorin proteins to the plasma membrane but does not primarily determine their localization in membrane microdomains. New Phytol 203:758–769
Lingwood D, Simons K (2010) Lipid rafts as a membrane-organizing principle. Science 327:46–50
Gronnier J, Crowet JM, Habenstein B, Nasir MN, Bayle V, Hosy E, Platre MP, Gouguet P, Raffaele S, Martinez D et al (2017) Structural basis for plant plasma membrane protein dynamics and organization into functional nanodomains. Elife 6:e26404
Ernst AM, Contreras FX, Brugger B, Wieland F (2010) Determinants of specificity at the protein-lipid interface in membranes. FEBS Lett 584:1713–1720
Szymanski WG, Zauber H, Erban A, Gorka M, Wu XN, Schulze WX (2015) Cytoskeletal components define protein location to membrane microdomains. Mol Cell Proteomics 14:2493–2509
Lv X, Jing Y, Xiao J, Zhang Y, Zhu Y, Julian R, Lin J (2017) Membrane microdomains and the cytoskeleton constrain AtHIR1 dynamics and facilitate the formation of an AtHIR1-associated immune complex. Plant J 90:3–16
Owen DM, Williamson D, Rentero C, Gaus K (2009) Quantitative microscopy: protein dynamics and membrane organisation. Traffic 10:962–971
Wang L, Li H, Lv X, Chen T, Li R, Xue Y, Jiang J, Jin B, Baluska F, Samaj J et al (2015) Spatiotemporal dynamics of the BRI1 receptor and its regulation by membrane microdomains in living Arabidopsis cells. Mol Plant 8:1334–1349
Haney CH, Long SR (2010) Plant Flotillins are required for infection by nitrogen-fixing bacteria. Proc Natl Acad Sci USA 107:478–483
Cui Y, Li X, Yu M, Li R, Fan L, Zhu Y, Lin J (2018) Sterols regulate endocytic pathways during flg22-induced defense responses in Arabidopsis. Development 145(19):dev165688
Bucherl CA, Jarsch IK, Schudoma C, Segonzac C, Mbengue M, Robatzek S, MacLean D, Ott T, Zipfel C (2017) Plant immune and growth receptors share common signalling components but localise to distinct plasma membrane nanodomains. Elife 6:e25114
Perraki A, Gronnier J, Gouguet P, Boudsocq M, Deroubaix AF, Simon V, German-Retana S, Legrand A, Habenstein B, Zipfel C et al (2018) REM1.3’s phospho-status defines its plasma membrane nanodomain organization and activity in restricting PVX cell-to-cell movement. PLoS Pathog 14:e1007378
Qi Y, Tsuda K, le Nguyen V, Wang X, Lin J, Murphy AS, Glazebrook J, Thordal-Christensen H, Katagiri F (2011) Physical association of Arabidopsis hypersensitive induced reaction proteins (HIRs) with the immune receptor RPS2. J Biol Chem 286:31297–31307
Ishikawa T, Aki T, Yanagisawa S, Uchimiya H, Kawai-Yamada M (2015) Overexpression of BAX INHIBITOR-1 links plasma membrane microdomain proteins to stress. Plant Physiol 169:1333–1343
Xue Y, Xing J, Wan Y, Lv X, Fan L, Zhang Y, Song K, Wang L, Wang X, Deng X et al (2018) Arabidopsis blue light receptor phototropin 1 undergoes blue light-induced activation in membrane microdomains. Mol Plant 11:846–859
Baral A, Irani NG, Fujimoto M, Nakano A, Mayor S, Mathew MK (2015) Salt-induced remodeling of spatially restricted clathrin-independent endocytic pathways in Arabidopsis root. Plant Cell 27:1297–1315
Claus LAN, Savatin DV, Russinova E (2018) The crossroads of receptor-mediated signaling and endocytosis in plants. J Integr Plant Biol 60:827–840
Mbengue M, Bourdais G, Gervasi F, Beck M, Zhou J, Spallek T, Bartels S, Boller T, Ueda T, Kuhn H et al (2016) Clathrin-dependent endocytosis is required for immunity mediated by pattern recognition receptor kinases. Proc Natl Acad Sci USA 113:11034–11039
Yoshinari A, Hosokawa T, Amano T, Beier MP, Kunieda T, Shimada T, Hara-Nishimura I, Naito S, Takano J (2019) Polar localization of the borate exporter BOR1 requires AP2-Dependent endocytosis. Plant Physiol 179:1569–1580
Kroumanova K, Kocourkova D, Danek M, Lamparova L, Pospichalova R, Malinska K, Krckova Z, Burketova L, Valentova O, Martinec J et al (2019) Characterisation of Arabidopsis Flotillins in response to stresses. Biol Plant 63:144–152
Wang Q, Zhao Y, Luo W, Li R, He Q, Fang X, Michele RD, Ast C, von Wiren N, Lin J (2013) Single-particle analysis reveals shutoff control of the Arabidopsis ammonium transporter AMT1;3 by clustering and internalization. Proc Natl Acad Sci USA 110:13204–13209
Slupianek A, Kasprowicz-Maluski A, Myskow E, Turzanska M, Sokolowska K (2019) Endocytosis acts as transport pathway in wood. New Phytol 222:1846–1861
Li X, Luu DT, Maurel C, Lin J (2013) Probing plasma membrane dynamics at the single-molecule level. Trends Plant Sci 18:617–624
Viola A, Gupta N (2007) Tether and trap: regulation of membrane-raft dynamics by actin-binding proteins. Nat Rev Immunol 7:889–896
Szymanski DB, Cosgrove DJ (2009) Dynamic coordination of cytoskeletal and cell wall systems during plant cell morphogenesis. Curr Biol 19:R800–R811
McKenna JF, Tolmie AF, Runions J (2014) Across the great divide: the plant cell surface continuum. Curr Opin Plant Biol 22:132–140
Martiniere A, Lavagi I, Nageswaran G, Rolfe DJ, Maneta-Peyret L, Luu DT, Botchway SW, Webb SE, Mongrand S, Maurel C et al (2012) Cell wall constrains lateral diffusion of plant plasma-membrane proteins. Proc Natl Acad Sci USA 109:12805–12810
Sorci-Thomas MG, Thomas MJ (2016) Microdomains, inflammation, and atherosclerosis. Circ Res 118:679–691
Goldfinger LE, Michael JV (2017) Regulation of Ras signaling and function by plasma membrane microdomains. Biosci Trends 11:23–40
Takahashi D, Kawamura Y, Uemura M (2013) Detergent-resistant plasma membrane proteome to elucidate microdomain functions in plant cells. Front Plant Sci 4:27
Willemsen V, Friml J, Grebe M, van den Toorn A, Palme K, Scheres B (2003) Cell polarity and PIN protein positioning in Arabidopsis require STEROL METHYLTRANSFERASE1 function. Plant Cell 15:612–625
Men S, Boutte Y, Ikeda Y, Li X, Palme K, Stierhof YD, Hartmann MA, Moritz T, Grebe M (2008) Sterol-dependent endocytosis mediates post-cytokinetic acquisition of PIN2 auxin efflux carrier polarity. Nat Cell Biol 10:237–244
Grison MS, Brocard L, Fouillen L, Nicolas W, Wewer V, Dormann P, Nacir H, Benitez-Alfonso Y, Claverol S, Germain V et al (2015) Specific membrane lipid composition is important for plasmodesmata function in Arabidopsis. Plant Cell 27:1228–1250
Zavaliev R, Dong X, Epel BL (2016) Glycosylphosphatidylinositol (GPI) modification serves as a primary plasmodesmal sorting signal. Plant Physiol 172:1061–1073
Raffaele S, Bayer E, Lafarge D, Cluzet S, German Retana S, Boubekeur T, Leborgne-Castel N, Carde JP, Lherminier J, Noirot E et al (2009) Remorin, a solanaceae protein resident in membrane rafts and plasmodesmata, impairs potato virus X movement. Plant Cell 21:1541–1555
Li S, Su X, Zhang B, Huang Q, Hu Z, Lu M (2013) Molecular cloning and functional analysis of the Populus deltoides remorin gene PdREM. Tree Physiol 33:1111–1121
Gui J, Liu C, Shen J, Li L (2014) Grain setting defect1, encoding a remorin protein, affects the grain setting in rice through regulating plasmodesmatal conductance. Plant Physiol 166:1463–1478
Colebatch G, Desbrosses G, Ott T, Krusell L, Montanari O, Kloska S, Kopka J, Udvardi MK (2004) Global changes in transcription orchestrate metabolic differentiation during symbiotic nitrogen fixation in Lotus japonicus. Plant J 39:487–512
El Yahyaoui F, Kuster H, Ben Amor B, Hohnjec N, Puhler A, Becker A, Gouzy J, Vernie T, Gough C, Niebel A et al (2004) Expression profiling in Medicago truncatula identifies more than 750 genes differentially expressed during nodulation, including many potential regulators of the symbiotic program. Plant Physiol 136:3159–3176
Coaker GL, Willard B, Kinter M, Stockinger EJ, Francis DM (2004) Proteomic analysis of resistance mediated by Rcm 2.0 and Rcm 5.1, two loci controlling resistance to bacterial canker of tomato. Mol Plant Microbe Interact 17:1019–1028
Lefebvre B, Timmers T, Mbengue M, Moreau S, Herve C, Toth K, Bittencourt-Silvestre J, Klaus D, Deslandes L, Godiard L et al (2010) A remorin protein interacts with symbiotic receptors and regulates bacterial infection. Proc Natl Acad Sci USA 107:2343–2348
Toth K, Stratil TF, Madsen EB, Ye J, Popp C, Antolin-Llovera M, Grossmann C, Jensen ON, Schussler A, Parniske M et al (2012) Functional domain analysis of the Remorin protein LjSYMREM1 in Lotus japonicus. PLoS One 7:e30817
Jamann TM, Luo X, Morales L, Kolkman JM, Chung CL, Nelson RJ (2016) A remorin gene is implicated in quantitative disease resistance in maize. Theor Appl Genet 129:591–602
Son S, Oh CJ, An CS (2014) Arabidopsis thaliana remorins interact with SnRK1 and play a role in susceptibility to beet curly top virus and beet severe curly top virus. Plant Pathol J 30:269–278
Liu J, Elmore JM, Fuglsang AT, Palmgren MG, Staskawicz BJ, Coaker G (2009) RIN4 functions with plasma membrane H+ -ATPases to regulate stomatal apertures during pathogen attack. PLoS Biol 7:e1000139
Jung HW, Lim CW, Lee SC, Choi HW, Hwang CH, Hwang BK (2008) Distinct roles of the pepper hypersensitive induced reaction protein gene CaHIR1 in disease and osmotic stress, as determined by comparative transcriptome and proteome analyses. Planta 227:409–425
Duan Y, Guo J, Shi X, Guan X, Liu F, Bai P, Huang L, Kang Z (2013) Wheat hypersensitive-induced reaction genes TaHIR1 and TaHIR3 are involved in response to stripe rust fungus infection and abiotic stresses. Plant Cell Rep 32:273–283
Jung HW, Hwang BK (2007) The leucine-rich repeat (LRR) protein, CaLRR1, interacts with the hypersensitive induced reaction (HIR) protein, CaHIR1, and suppresses cell death induced by the CaHIR1 protein. Mol Plant Pathol 8:503–514
Zhou L, Cheung MY, Zhang Q, Lei CL, Zhang SH, Sun SS, Lam HM (2009) A novel simple extracellular leucine-rich repeat (eLRR) domain protein from rice (OsLRR1) enters the endosomal pathway and interacts with the hypersensitive-induced reaction protein 1 (OsHIR1). Plant Cell Environ 32:1804–1820
Checker VG, Khurana P (2013) Molecular and functional characterization of mulberry EST encoding remorin (MiREM) involved in abiotic stress. Plant Cell Rep 32:1729–1741
Yue J, Li C, Liu Y, Yu J (2014) A remorin gene SiREM6, the target gene of SiARDP, from foxtail millet (Setaria italica) promotes high salt tolerance in transgenic Arabidopsis. PLoS One 9:e100772
Cui Y, Zhang X, Yu M, Zhu Y, Xing J, Lin J (2019) Techniques for detecting protein-protein interactions in living cells: principles, limitations, and recent progress. Sci China Life Sci 62:619–632
Acknowledgements
We thank lab members for helpful discussions and critical reading of the paper. This work was supported by the State ‘13.5’ Key Research Program of China (No. 2016YFD0600102), the National Natural Science Foundation of China (31530084, 31761133009, 31670182, 31401149), and the Fundamental Research Funds for the Central Universities (2019ZY29, 2017ZY10) and the Program of Introducing Talents of Discipline to Universities (111 project, B13007).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Yu, M., Cui, Y., Zhang, X. et al. Organization and dynamics of functional plant membrane microdomains. Cell. Mol. Life Sci. 77, 275–287 (2020). https://doi.org/10.1007/s00018-019-03270-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-019-03270-7