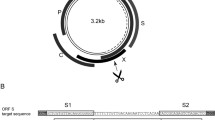
Abstract
Covalently closed circular DNA (cccDNA) of hepatitis B virus (HBV) is the major cause of viral persistence and chronic hepatitis B. CRISPR/Cas9 nucleases can specifically target HBV cccDNA for decay, but off-target effects of nucleases in the human genome limit their clinical utility. CRISPR/Cas9 systems from four different species were co-expressed in cell lines with guide RNAs targeting conserved regions of the HBV genome. CRISPR/Cas9 systems from Streptococcus pyogenes (Sp) and Streptococcus thermophilus (St) targeting conserved regions of the HBV genome blocked HBV replication and, most importantly, resulted in degradation of over 90% of HBV cccDNA by 6 days post-transfection. Degradation of HBV cccDNA was impaired by inhibition of non-homologous end-joining pathway and resulted in an erroneous repair of HBV cccDNA. HBV cccDNA methylation also affected antiviral activity of CRISPR/Cas9. Single-nucleotide HBV genetic variants did not impact anti-HBV activity of St CRISPR/Cas9, suggesting its utility in targeting many HBV variants. However, two or more mismatches impaired or blocked CRISPR/Cas9 activity, indicating that host DNA will not likely be targeted. Deep sequencing revealed that Sp CRISPR/Cas9 induced off-target mutagenesis, whereas St CRISPR/Cas9 had no effect on the host genome. St CRISPR/Cas9 system represents the safest system with high anti-HBV activity.







Similar content being viewed by others
References
Schweitzer A, Horn J, Mikolajczyk RT, Krause G, Ott JJ (2015) Estimations of worldwide prevalence of chronic hepatitis B virus infection: a systematic review of data published between 1965 and 2013. Lancet 386(10003):1546–1555
World Health Organization (2017) Global hepatitis report 2017. World Health Organization, Geneva
Yue D et al (2016) Hepatitis B virus X protein (HBx)-induced abnormalities of nucleic acid metabolism revealed by 1 H-NMR-based metabonomics. Sci Rep 6:24430
Li J et al (2017) Hepatitis B virus X protein inhibits apoptosis by modulating endoplasmic reticulum stress response. Oncotarget 8(56):96027–96034
Matsuda Y et al (2013) DNA damage sensor γ-H2AX is increased in preneoplastic lesions of hepatocellular carcinoma. Sci World J 2013:597095
Guerrieri F et al (2017) Genome-wide identification of direct HBx genomic targets. BMC Genom 18(1):184
Lamontagne RJ, Bagga S, Bouchard MJ (2016) Hepatitis B virus molecular biology and pathogenesis. Hepatoma Res 2:163
Allweiss L, Dandri M (2017) The role of cccDNA in HBV maintenance. Viruses 9(6):156
Koumbi L (2015) Current and future antiviral drug therapies of hepatitis B chronic infection. World J Hepatol 7(8):1030–1040
Chulanov VP, Zueva AP, Kostyushev DS, Brezgin SA, Volchkova EV, Maleyev VV (2017) Hepatitis C can be cured: will hepatitis B become next? Ter Arkh 89(11):4–13
Kostyusheva A, Kostyushev D, Brezgin S, Volchkova E, Chulanov V (2018) Clinical implications of hepatitis B virus RNA and covalently closed circular DNA in monitoring patients with chronic hepatitis B today with a gaze into the future: the field is unprepared for a sterilizing cure. Genes 9:10
Alter H et al (2018) A research agenda for curing chronic hepatitis B virus infection. Hepatology 67(3):1127–1131
Jiang F, Doudna JA (2017) CRISPR–Cas9 structures and mechanisms. Annu Rev Biophys 46(March):505–529
Mali P et al (2013) RNA-guided human genome engineering via Cas9. Science (80-.) 339(6121):823–826
Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA (2014) DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507(7490):62–67
Kleinstiver BP et al (2016) High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529(7587):490–495
Lee JK et al (2018) Directed evolution of CRISPR–Cas9 to increase its specificity. Nat Commun 9(1):3048
Tycko J, Myer VE, Hsu PD (2016) Methods for optimizing CRISPR–Cas9 genome editing specificity. Mol Cell 63(3):355–370
Lee CM, Cradick TJ, Bao G (2016) The Neisseria meningitidis CRISPR–Cas9 system enables specific genome editing in mammalian cells. Mol Ther 24(3):645–654
Müller M et al (2016) Streptococcus thermophilus CRISPR–Cas9 systems enable specific editing of the human genome. Mol Ther 24(3):636–644
Ran FA et al (2015) In vivo genome editing using Staphylococcus aureus Cas9. Nature 520(7546):186–191
Liu Y et al (2018) Inhibition of hepatitis B virus replication via HBV DNA cleavage by Cas9 from Staphylococcus aureus. Antiviral Res 152:58–67
Scott T et al (2017) ssAAVs containing cassettes encoding SaCas9 and guides targeting hepatitis B virus inactivate replication of the virus in cultured cells. Sci Rep 7(1):7401
Price AA, Sampson TR, Ratner HK, Grakoui A, Weiss DS (2015) Cas9-mediated targeting of viral RNA in eukaryotic cells. Proc Natl Acad Sci 112(19):6164–6169
Croagh CMN, Desmond PV, Bell SJ (2015) Genotypes and viral variants in chronic hepatitis B: a review of epidemiology and clinical relevance. World J Hepatol 7(3):289
Liu X, Hao R, Chen S, Guo D, Chen Y (2015) Inhibition of hepatitis B virus by the CRISPR/Cas9 system via targeting the conserved regions of the viral genome. J Gen Virol 96(8):2252–2261
Cong L et al (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339(6121):819–823
Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA (2013) RNA-guided editing of bacterial genomes using CRISPR–Cas systems. Nat Biotechnol 31(3):233–239
Ramanan V et al (2015) CRISPR/Cas9 cleavage of viral DNA efficiently suppresses hepatitis B virus. Sci Rep 5:10833
Stemmer M, Thumberger T, del Sol Keyer M, Wittbrodt J, Mateo JL (2015) CCTop: an intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS One 10(4):e0124633
Labuhn M et al (2017) Refined sgRNA efficacy prediction improves large-and small-scale CRISPR–Cas9 applications. Nucleic Acids Res 46(3):1375–1385
Yang H-C, Kao J-H (2014) Persistence of hepatitis B virus covalently closed circular DNA in hepatocytes: molecular mechanisms and clinical significance. Emerg Microbes Infect 3(9):e64
Li G et al (2018) Recombinant covalently closed circular DNA of hepatitis B virus induces long-term viral persistence with chronic hepatitis in a mouse model. Hepatology 67(1):56–70
Guo X et al (2016) The recombined cccDNA produced using minicircle technology mimicked HBV genome in structure and function closely. Sci Rep 6:25552
Hirano H et al (2016) Structure and Engineering of Francisella novicida Cas9. Cell 164(5):950–961
Daer R, Barrett CM, Haynes KA (2017) Histone modifications and active gene expression are associated with enhanced CRISPR activity in de-silenced chromatin. bioRxiv 228601. https://doi.org/10.1101/228601
Chen X, Rinsma M, Janssen JM, Liu J, Maggio I, Gonçalves MAFV (2016) Probing the impact of chromatin conformation on genome editing tools. Nucleic Acids Res 44(13):6482–6492
Valton J et al (2012) Overcoming TALE DNA binding domain sensitivity to cytosine methylation. J Biol Chem 287(46):38427–38432
Hsu PD et al (2013) DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 31(9):827–832
Yarrington RM, Verma S, Schwartz S, Trautman JK, Carroll D (2018) Nucleosomes inhibit target cleavage by CRISPR–Cas9 in vivo. Proc Natl Acad Sci USA 115:9351
Tropberger P, Mercier A, Robinson M, Zhong W, Ganem DE, Holdorf M (2015) Mapping of histone modifications in episomal HBV cccDNA uncovers an unusual chromatin organization amenable to epigenetic manipulation. Proc Natl Acad Sci USA 112(42):E5715–E5724
Lucifora J et al (2014) Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science (80-.) 343(6176):1221–1228
Kostyushev DS et al (2017) Overexpression of DNA-methyltransferases in persistency of cccDNA pool in chronic hepatitis B. Ter Arkh 89(11):21–26
Zhang Y et al (2013) Comparative analysis of CpG Islands among HBV genotypes. PLoS One 8(2):1–8
Niazi MT et al (2014) Effects of dna-dependent protein kinase inhibition by NU7026 on DNA repair and cell survival in irradiated gastric cancer cell line N87. Curr Oncol 21(2):91–96
Caligiuri P, Cerruti R, Icardi G, Bruzzone B (2016) Overview of hepatitis B virus mutations and their implications in the management of infection. World J Gastroenterol 22(1):145
Kim JH, Park YK, Park E-S, Kim K-H (2014) Molecular diagnosis and treatment of drug-resistant hepatitis B virus. World J Gastroenterol 20(19):5708
Köck J, Blum HE (2008) Hypermutation of hepatitis B virus genomes by APOBEC3G, APOBEC3C and APOBEC3H. J Gen Virol 89(5):1184–1191
Vartanian JP et al (2010) Massive APOBEC3 editing of hepatitis B viral DNA in cirrhosis. PLoS Pathog 6(5):1–9
Noguchi C et al (2005) G to A hypermutation of hepatitis B virus. Hepatology 41(3):626–633
Li F et al (2015) Whole genome characterization of hepatitis B virus quasispecies with massively parallel pyrosequencing. Clin Microbiol Infect 21(3):280–287
Liu Y et al (2015) Hepatitis B virus polymerase disrupts K63-linked ubiquitination of STING to block innate cytosolic DNA-sensing pathways. J Virol 89(4):2287–2300
Aragri M et al (2016) Multiple hepatitis B virus (HBV) quasispecies and immune-escape mutations are present in HBV surface antigen and reverse transcriptase of patients with acute hepatitis B. J Infect Dis 213(12):1897–1905
Wang J et al (2015) Dual gRNAs guided CRISPR/Cas9 system inhibits hepatitis B virus replication. World J Gastroenterol 21(32):9554–9565
Zhang XH, Tee LY, Wang XG, Huang QS, Yang SH (2015) Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol Ther Nucleic Acids 4:11
Cho SW et al (2014) Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res 24(1):132–141
Fu Y et al (2013) High-frequency off-target mutagenesis induced by CRISPR–Cas nucleases in human cells. Nat Biotech 31(9):822–826
Kosicki M, Bradley A (2018) Repair of CRISPR–Cas9-induced double-stranded breaks leads to large deletions and complex rearrangements. Nat Biotechnol 36:765–771
Akcakaya P et al. (2018) In vivo CRISPR editing with no detectable genome-wide off-target mutations. Nature 561:416–419
Esvelt KM, Mali P, Braff JL, Moosburner M, Yaung SJ, Church GM (2013) Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods 10(11):1116–1123
Zhu W et al (2016) CRISPR/Cas9 produces anti-hepatitis B virus effect in hepatoma cells and transgenic mouse. Virus Res 217:125–132
Seeger C, Sohn JA (2016) Complete spectrum of CRISPR/Cas9-induced mutations on HBV cccDNA. Mol Ther 24(7):1258–1266
Dong C, Qu L, Wang H, Wei L, Dong Y, Xiong S (2015) Targeting hepatitis B virus cccDNA by CRISPR/Cas9 nuclease efficiently inhibits viral replication. Antiviral Res 118:110–117
Kennedy EM, Kornepati AVR, Cullen BR (2015) Targeting hepatitis B virus cccDNA using CRISPR/Cas9. Antiviral Res 123:188–192
Ng H, Dean N (2017) Dramatic improvement of CRISPR/Cas9 editing in Candida albicans by increased single guide RNA expression. mSphere 2(2):e00385–e00416
Yuen G et al (2017) CRISPR/Cas9-mediated gene knockout is insensitive to target copy number but is dependent on guide RNA potency and Cas9/sgRNA threshold expression level. Nucleic Acids Res 45(20):12039–12053
Hemmi H et al (2000) A Toll-like receptor recognizes bacterial DNA. Nature 408(6813):740–745
Ishii KJ et al (2006) A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat Immunol 7(1):40–48
Stetson DB, Medzhitov R (2006) Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity 24(1):93–103
Ridruejo E et al (2014) Relapse rates in chronic hepatitis B naive patients after discontinuation of antiviral therapy with entecavir. J Viral Hepatol 21(8):590–596
Hoofnagle JH (2009) Reactivation of hepatitis B. Hepatology 49(5 Suppl):S156–S165
Hong X, Kim ES, Guo H (2017) Epigenetic regulation of hepatitis B virus covalently closed circular DNA: implications for epigenetic therapy against chronic hepatitis B. Hepatology 66(6):2066–2077
Jain S et al (2015) Comprehensive DNA methylation analysis of hepatitis B virus genome in infected liver tissues. Sci Rep 5:10478
Vivekanandan P, Daniel HD-J, Kannangai R, Martinez-Murillo F, Torbenson M (2010) Hepatitis B virus replication induces methylation of both host and viral DNA. J Virol 84(9):4321–4329
Zhang Y et al (2014) Transcription of hepatitis B virus covalently closed circular DNA is regulated by CpG methylation during chronic infection. PLoS One 9(10):e110442
Koumbi L, Karayiannis P (2016) The epigenetic control of hepatitis B virus modulates the outcome of infection. Front Microbiol 6(JAN):1–9
Kaur P et al (2010) DNA methylation of hepatitis B virus (HBV) genome associated with the development of hepatocellular carcinoma and occult HBV infection. J Infect Dis 202(5):700–704
Cai D, Nie H, Yan R, Guo J-T, Block TM, Guo H (2013) A southern blot assay for detection of hepatitis B virus covalently closed circular DNA from cell cultures. Methods Mol Biol 1030:151–161
Acknowledgements
We thank Konstantin Severinov and Dieter Glebe for their helpful contributions, Yurii Babin and Konstantin Flyagin for technical assistance, and Vladimir Simirskii for access to microscopy.
Author information
Authors and Affiliations
Contributions
DK, SB, and AK conducted all experiments; DZ and SB generated recombinant cccDNA and created gRNAs; DZ analyzed off-target sites and designed specific primers; IG conducted sequencing; DK conceived the project; AK helped conceive experiments with mutant gRNAs; DK, DZ, SB, and AK processed the data; DK wrote the manuscript; VC guided the study and revised the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no potential conflicts of interests. The authors have applied for patents concerning the use of Cas9 proteins and gRNAs for HBV therapy.
Funding
This work was supported by RSF Grant no. 16-15-10426.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kostyushev, D., Brezgin, S., Kostyusheva, A. et al. Orthologous CRISPR/Cas9 systems for specific and efficient degradation of covalently closed circular DNA of hepatitis B virus. Cell. Mol. Life Sci. 76, 1779–1794 (2019). https://doi.org/10.1007/s00018-019-03021-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-019-03021-8