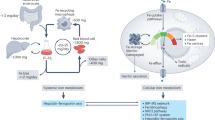
Abstract
Iron homeostasis is maintained at the cellular and systemic levels to assure adequate iron supply while preventing iron overload. The identification of genes mutated in patients with iron-related disorders or animal models with imbalances of iron homeostasis gave insight into the molecular mechanisms underlying processes critical for balancing iron levels, such as iron uptake, storage, export, and monitoring of available iron. MicroRNAs control genes involved in some of these processes adding an additional level of complexity to the regulation of iron metabolism. This review summarizes recent advances how miRNAs regulate iron homeostasis.


Similar content being viewed by others
References
Kim YK, Kim VN (2007) Processing of intronic microRNAs. EMBO J 26:775–783
Saini HK, Griffiths-Jones S, Enright AJ (2007) Genomic analysis of human microRNA transcripts. Proc Natl Acad Sci USA 104:17719–17724
Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN (2004) MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23:4051–4060
Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ (2004) Processing of primary microRNAs by the Microprocessor complex. Nature 432:231–235
Griffiths-Jones S, Saini HK, van DS, Enright AJ (2008) miRBase: tools for microRNA genomics. Nucleic Acids Res 36:D154–D158
Sun W, Julie Li YS, Huang HD, Shyy JY, Chien S (2010) microRNA: a master regulator of cellular processes for bioengineering systems 1. Annu Rev Biomed Eng 12:1–27
Bushati N, Cohen SM (2008) MicroRNAs in neurodegeneration 2. Curr Opin Neurobiol 18:292–296
Davis BN, Hilyard AC, Nguyen PH, Lagna G, Hata A (2010) Smad proteins bind a conserved RNA sequence to promote microRNA maturation by Drosha. Mol Cell 39:373–384
Davis BN, Hilyard AC, Lagna G, Hata A (2008) SMAD proteins control DROSHA-mediated microRNA maturation. Nature 454:56–61
Bartley AN, Yao H, Barkoh BA, Ivan C, Mishra BM, Rashid A, Calin GA, Luthra R, Hamilton SR (2011) Complex patterns of altered MicroRNA expression during the adenoma-adenocarcinoma sequence for microsatellite-stable colorectal cancer. Clin Cancer Res 17:7283–7293
Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB (2003) Prediction of mammalian microRNA targets 2. Cell 115:787–798
Karreth FA, Tay Y, Perna D, Ala U, Tan SM, Rust AG, DeNicola G, Webster KA, Weiss D, Perez-Mancera PA et al (2011) In vivo identification of tumor- suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell 147:382–395
Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, Karreth F, Poliseno L, Provero P, Di CF et al (2011) Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell 147:344–357
Sumazin P, Yang X, Chiu HS, Chung WJ, Iyer A, Llobet-Navas D, Rajbhandari P, Bansal M, Guarnieri P, Silva J et al (2011) An extensive microRNA-mediated network of RNA–RNA interactions regulates established oncogenic pathways in glioblastoma. Cell 147:370–381
Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP (2010) A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 465:1033–1038
Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I (2011) A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147:358–369
Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP (2011) A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146:353–358
Corcoran DL, Pandit KV, Gordon B, Bhattacharjee A, Kaminski N, Benos PV (2009) Features of mammalian microRNA promoters emerge from polymerase II chromatin immunoprecipitation data. PLoS One 4:e5279
Shiohama A, Sasaki T, Noda S, Minoshima S, Shimizu N (2003) Molecular cloning and expression analysis of a novel gene DGCR8 located in the DiGeorge syndrome chromosomal region. Biochem Biophys Res Commun 304:184–190
Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R (2004) The Microprocessor complex mediates the genesis of microRNAs. Nature 432:235–240
Gregory RI, Chendrimada TP, Shiekhattar R (2006) MicroRNA biogenesis: isolation and characterization of the microprocessor complex. Methods Mol Biol 342:33–47
Krol J, Loedige I, Filipowicz W (2010) The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 11:597–610
Faller M, Matsunaga M, Yin S, Loo JA, Guo F (2007) Heme is involved in microRNA processing. Nat Struct Mol Biol 14:23–29
Barr I, Smith AT, Senturia R, Chen Y, Scheidemantle BD, Burstyn JN, Guo F (2011) DiGeorge critical region 8 (DGCR8) is a double-cysteine-ligated heme protein. J Biol Chem 286:16716–16725
Faller M, Toso D, Matsunaga M, Atanasov I, Senturia R, Chen Y, Zhou ZH, Guo F (2010) DGCR8 recognizes primary transcripts of microRNAs through highly cooperative binding and formation of higher-order structures 1. RNA 16:1570–1583
Senturia R, Faller M, Yin S, Loo JA, Cascio D, Sawaya MR, Hwang D, Clubb RT, Guo F (2010) Structure of the dimerization domain of DiGeorge critical region 8 2. Protein Sci 19:1354–1365
Barr I, Smith AT, Chen Y, Senturia R, Burstyn JN, Guo F (2012) Ferric, not ferrous, heme activates RNA-binding protein DGCR8 for primary microRNA processing. Proc Natl Acad Sci USA 109:1919–1924
Krause A, Neitz S, Magert HJ, Schulz A, Forssmann WG, Schulz-Knappe P, Adermann K (2000) LEAP-1, a novel highly disulfide-bonded human peptide, exhibits antimicrobial activity. FEBS Lett 480:147–150
Park CH, Valore EV, Waring AJ, Ganz T (2001) Hepcidin, a urinary antimicrobial peptide synthesized in the liver. J Biol Chem 276:7806–7810
Ward DM, Kaplan J (2012) Ferroportin-mediated iron transport: expression and regulation. Biochim Biophys Acta 25:119–131
Nemeth E, Tuttle MS, Powelson J, Vaughn MB, Donovan A, Ward DM, Ganz T, Kaplan J (2004) Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science 306:2090–2093
Muckenthaler M, Roy CN, Custodio AO, Minana B, deGraaf J, Montross LK, Andrews NC, Hentze MW (2003) Regulatory defects in liver and intestine implicate abnormal hepcidin and Cybrd1 expression in mouse hemochromatosis. Nat Genet 34:102–107
Bridle KR, Frazer DM, Wilkins SJ, Dixon JL, Purdie DM, Crawford DH, Subramaniam VN, Powell LW, Anderson GJ, Ramm GA (2003) Disrupted hepcidin regulation in HFE-associated haemochromatosis and the liver as a regulator of body iron homoeostasis 1. Lancet 361:669–673
Papanikolaou G, Samuels ME, Ludwig EH, MacDonald ML, Franchini PL, Dube MP, Andres L, MacFarlane J, Sakellaropoulos N, Politou M et al (2004) Mutations in HFE2 cause iron overload in chromosome 1q-linked juvenile hemochromatosis 1. Nat Genet 36:77–82
Huang FW, Pinkus JL, Pinkus GS, Fleming MD, Andrews NC (2005) A mouse model of juvenile hemochromatosis 1. J Clin Invest 115:2187–2191
Niederkofler V, Salie R, Arber S (2005) Hemojuvelin is essential for dietary iron sensing, and its mutation leads to severe iron overload 1. J Clin Invest 115:2180–2186
Camaschella C, Roetto A, Cali A, De GM, Garozzo G, Carella M, Majorano N, Totaro A, Gasparini P (2000) The gene TFR2 is mutated in a new type of haemochromatosis mapping to 7q22 1. Nat Genet 25:14–15
Fleming RE, Ahmann JR, Migas MC, Waheed A, Koeffler HP, Kawabata H, Britton RS, Bacon BR, Sly WS (2002) Targeted mutagenesis of the murine transferrin receptor-2 gene produces hemochromatosis 1. Proc Natl Acad Sci USA 99:10653–10658
Silvestri L, Pagani A, Nai A, De DI, Kaplan J, Camaschella C (2008) The serine protease matriptase-2 (TMPRSS6) inhibits hepcidin activation by cleaving membrane hemojuvelin 1. Cell Metab 8:502–511
Mleczko-Sanecka K, Casanovas G, Ragab A, Breitkopf K, Muller A, Boutros M, Dooley S, Hentze MW, Muckenthaler MU (2010) SMAD7 controls iron metabolism as a potent inhibitor of hepcidin expression 1. Blood 115:2657–2665
Muckenthaler MU, Galy B, Hentze MW (2008) Systemic iron homeostasis and the iron-responsive element/iron-regulatory protein (IRE/IRP) regulatory network. Annu Rev Nutr 28:197–213
Sanchez M, Galy B, Schwanhaeusser B, Blake J, Bahr-Ivacevic T, Benes V, Selbach M, Muckenthaler MU, Hentze MW (2011) Iron regulatory protein-1 and -2: transcriptome-wide definition of binding mRNAs and shaping of the cellular proteome by iron regulatory proteins. Blood 118:e168–e179
Smith SR, Cooperman S, Lavaute T, Tresser N, Ghosh M, Meyron-Holtz E, Land W, Ollivierre H, Jortner B, Switzer R III et al (2004) Severity of neurodegeneration correlates with compromise of iron metabolism in mice with iron regulatory protein deficiencies 1. Ann NY Acad Sci 1012:65–83
Ohgami RS, Campagna DR, Greer EL, Antiochos B, McDonald A, Chen J, Sharp JJ, Fujiwara Y, Barker JE, Fleming MD (2005) Identification of a ferrireductase required for efficient transferrin-dependent iron uptake in erythroid cells 1. Nat Genet 37:1264–1269
Magro G, Cataldo I, Amico P, Torrisi A, Vecchio GM, Parenti R, Asioli S, Recupero D, D’Agata V, Mucignat MT et al (2011) Aberrant expression of TfR1/CD71 in thyroid carcinomas identifies a novel potential diagnostic marker and therapeutic target 1. Thyroid 21:267–277
Kwok JC, Richardson DR (2002) The iron metabolism of neoplastic cells: alterations that facilitate proliferation? 1. Crit Rev Oncol Hematol 42:65–78
Rovera G, Santoli D, Damsky C (1979) Human promyelocytic leukemia cells in culture differentiate into macrophage-like cells when treated with a phorbol diester 1. Proc Natl Acad Sci USA 76:2779–2783
Schaar DG, Medina DJ, Moore DF, Strair RK, Ting Y (2009) miR-320 targets transferrin receptor 1 (CD71) and inhibits cell proliferation 1. Exp Hematol 37:245–255
Hubert N, Hentze MW (2002) Previously uncharacterized isoforms of divalent metal transporter (DMT)-1: implications for regulation and cellular function 1. Proc Natl Acad Sci USA 99:12345–12350
Gunshin H, Allerson CR, Polycarpou-Schwarz M, Rofts A, Rogers JT, Kishi F, Hentze MW, Rouault TA, Andrews NC, Hediger MA (2001) Iron-dependent regulation of the divalent metal ion transporter 1. FEBS Lett 509:309–316
Tchernitchko D, Bourgeois M, Martin ME, Beaumont C (2002) Expression of the two mRNA isoforms of the iron transporter Nramp2/DMTI in mice and function of the iron responsive element 1. Biochem J 363:449–455
Canonne-Hergaux F, Zhang AS, Ponka P, Gros P (2001) Characterization of the iron transporter DMT1 (NRAMP2/DCT1) in red blood cells of normal and anemic mk/mk mice 1. Blood 98:3823–3830
Andolfo I, De FL, Asci R, Russo R, Colucci S, Gorrese M, Zollo M, Iolascon A (2010) Regulation of divalent metal transporter 1 (DMT1) non-IRE isoform by the microRNA Let-7d in erythroid cells 1. Haematologica 95:1244–1252
Chua AC, Olynyk JK, Leedman PJ, Trinder D (2004) Nontransferrin-bound iron uptake by hepatocytes is increased in the Hfe knockout mouse model of hereditary hemochromatosis. Blood 104:1519–1525
Mastrogiannaki M, Matak P, Keith B, Simon MC, Vaulont S, Peyssonnaux C (2009) HIF-2alpha, but not HIF-1alpha, promotes iron absorption in mice 1. J Clin Invest 119:1159–1166
Yeh KY, Yeh M, Polk P, Glass J (2011) Hypoxia-inducible factor-2alpha and iron absorptive gene expression in Belgrade rat intestine 2. Am J Physiol Gastrointest Liver Physiol 301:G82–G90
Shpyleva SI, Tryndyak VP, Kovalchuk O, Starlard-Davenport A, Chekhun VF, Beland FA, Pogribny IP (2011) Role of ferritin alterations in human breast cancer cells 2. Breast Cancer Res Treat 126:63–71
Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, Diehn M, Liu H, Panula SP, Chiao E et al (2009) Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells 1. Cell 138:592–603
Dykxhoorn DM, Wu Y, Xie H, Yu F, Lal A, Petrocca F, Martinvalet D, Song E, Lim B, Lieberman J (2009) miR-200 enhances mouse breast cancer cell colonization to form distant metastases 1. PLoS One 4:e7181
Castoldi M, Vujic SM, Altamura S, Elmen J, Lindow M, Kiss J, Stolte J, Sparla R, D’Alessandro LA, Klingmuller U et al (2011) The liver-specific microRNA miR-122 controls systemic iron homeostasis in mice. J Clin Invest 121:1386–1396
Bolondi G, Garuti C, Corradini E, Zoller H, Vogel W, Finkenstedt A, Babitt JL, Lin HY, Pietrangelo A (2010) Altered hepatic BMP signaling pathway in human HFE hemochromatosis 1. Blood Cells Mol Dis 45:308–312
Padgett KA, Lan RY, Leung PC, Lleo A, Dawson K, Pfeiff J, Mao TK, Coppel RL, Ansari AA, Gershwin ME (2009) Primary biliary cirrhosis is associated with altered hepatic microRNA expression 1. J Autoimmun 32:246–253
Kutay H, Bai S, Datta J, Motiwala T, Pogribny I, Frankel W, Jacob ST, Ghoshal K (2006) Downregulation of miR-122 in the rodent and human hepatocellular carcinomas 1. J Cell Biochem 99:671–678
Girard M, Jacquemin E, Munnich A, Lyonnet S, Henrion-Caude A (2008) miR-122, a paradigm for the role of microRNAs in the liver 1. J Hepatol 48:648–656
Kew MC (2009) Hepatic iron overload and hepatocellular carcinoma. Cancer Lett 286:38–43
Elmen J, Lindow M, Silahtaroglu A, Bak M, Christensen M, Lind-Thomsen A, Hedtjarn M, Hansen JB, Hansen HF, Straarup EM et al (2008) Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver 17. Nucleic Acids Res 36:1153–1162
Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R et al (2006) miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting 18. Cell Metab 3:87–98
Jopling CL, Norman KL, Sarnow P (2006) Positive and negative modulation of viral and cellular mRNAs by liver-specific microRNA miR-122 1. Cold Spring Harb Symp Quant Biol 71:369–376
Jopling CL (2008) Regulation of hepatitis C virus by microRNA-122 1. Biochem Soc Trans 36:1220–1223
Sarasin-Filipowicz M (2010) Interferon therapy of hepatitis C: molecular insights into success and failure 1. Swiss Med Wkly 140:3–11
Ryan JD, Altamura S, Devitt E, Mullins S, Lawless MW, Muckenthaler MU, Crowe J (2012) Pegylated interferon-alpha induced hypoferremia is associated with the immediate response to treatment in hepatitis C. Hepatology. doi:10.1002/hep.25666
Svasti S, Masaki S, Penglong T, Abe Y, Winichagoon P, Fucharoen S, Umemura T (2010) Expression of microRNA-451 in normal and thalassemic erythropoiesis. Ann Hematol 89:953–958
Geekiyanage H, Chan C (2011) MicroRNA-137/181c regulates serine palmitoyltransferase and in turn amyloid beta, novel targets in sporadic Alzheimer’s disease. J Neurosci 31:14820–14830
Yoon D, Pastore YD, Divoky V, Liu E, Mlodnicka AE, Rainey K, Ponka P, Semenza GL, Schumacher A, Prchal JT (2006) Hypoxia-inducible factor-1 deficiency results in dysregulated erythropoiesis signaling and iron homeostasis in mouse development 1. J Biol Chem 281:25703–25711
Favaro E, Ramachandran A, McCormick R, Gee H, Blancher C, Crosby M, Devlin C, Blick C, Buffa F, Li JL et al (2010) MicroRNA-210 regulates mitochondrial free radical response to hypoxia and krebs cycle in cancer cells by targeting iron sulfur cluster protein ISCU 4. PLoS One 5:e10345
Chan SY, Zhang YY, Hemann C, Mahoney CE, Zweier JL, Loscalzo J (2009) MicroRNA-210 controls mitochondrial metabolism during hypoxia by repressing the iron-sulfur cluster assembly proteins ISCU1/2 6. Cell Metab 10:273–284
Arosio P, Levi S (2010) Cytosolic and mitochondrial ferritins in the regulation of cellular iron homeostasis and oxidative damage. Biochim Biophys Acta 1800:783–792
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Castoldi, M., Muckenthaler, M.U. Regulation of iron homeostasis by microRNAs. Cell. Mol. Life Sci. 69, 3945–3952 (2012). https://doi.org/10.1007/s00018-012-1031-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-012-1031-4