Abstract
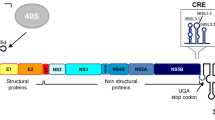
Hepatitis C virus (HCV) translation is mediated by an internal ribosome entry site (IRES) located at the 5′ end of the genomic RNA. The 3′ untranslatable region (3′UTR) stimulates translation by the recruitment of protein factors that simultaneously bind to the 5′ end of the viral genome. This leads to the formation of a macromolecular complex with a closed loop conformation, similar to that described for the cap-translated mRNAs. We previously demonstrated the existence of a long-range RNA–RNA interaction involving subdomain IIId of the IRES region and the stem–loop 5BSL3.2 of the CRE element at the 3′ end of the viral genome. The present study provides evidence that the enhancement of HCV IRES-dependent translation mediated by the 3′UTR is negatively controlled by the CRE region in the human hepatoma cell lines Huh-7 and Hep-G2 in a time-dependent manner. Domain 5BSL3.2 is the major partner in this process. Mutations in this motif lead to an increase in IRES activity by up to eightfold. These data support the existence of a functional high order structure in the HCV genome that involves two evolutionarily conserved RNA elements, domain IIId in the IRES and stem–loop 5BSL3.2 in the CRE region. This interaction could have a role in the circularisation of the viral genome.






Similar content being viewed by others
Abbreviations
- HCV:
-
Hepatitis C virus
- IRES:
-
Internal ribosome entry site
- UTR:
-
Untranslatable region
- CRE:
-
cis-acting replicating element
References
Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M (1989) Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359–362
Kuo G, Choo QL, Alter HJ et al (1989) An assay for circulating antibodies to a major etiologic virus of human non-A, non-B hepatitis. Science 244:362–364
Hoofnagle JH (1997) Hepatitis C: the clinical spectrum of disease. Hepatology 26:15S–20S
Kato N, Hijikata M, Ootsuyama Y, Nakagawa M, Ohkoshi S, Sugimura T, Shimotohno K (1990) Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc Natl Acad Sci USA 87:9524–9528
Takamizawa A, Mori C, Fuke I, Manabe S, Murakami S, Fujita J, Onishi E, Andoh T, Yoshida I, Okayama H (1991) Structure and organization of the hepatitis C virus genome isolated from human carriers. J Virol 65:1105–1113
Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A (1992) Internal ribosome entry site within hepatitis C virus RNA. J Virol 66:1476–1483
Wang C, Sarnow P, Siddiqui A (1993) Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J Virol 67:3338–3344
Kolykhalov AA, Mihalik K, Feinstone SM, Rice CM (2000) Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3’ nontranslated region are essential for virus replication in vivo. J Virol 74:2046–2051
Friebe P, Bartenschlager R (2002) Genetic analysis of sequences in the 3’ nontranslated region of hepatitis C virus that are important for RNA replication. J Virol 76:5326–5338
Yi M, Lemon SM (2003) 3’ nontranslated RNA signals required for replication of hepatitis C virus RNA. J Virol 77:3557–3568
Yi M, Lemon SM (2003) Structure-function analysis of the 3’ stem–loop of hepatitis C virus genomic RNA and its role in viral RNA replication. RNA 9:331–345
Song Y, Friebe P, Tzima E, Junemann C, Bartenschlager R, Niepmann M (2006) The hepatitis C virus RNA 3’-untranslated region strongly enhances translation directed by the internal ribosome entry site. J Virol 80:11579–11588
Reynolds JE, Kaminski A, Kettinen HJ, Grace K, Clarke BE, Carroll AR, Rowlands DJ, Jackson RJ (1995) Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J 14:6010–6020
Wang TH, Rijnbrand RC, Lemon SM (2000) Core protein-coding sequence, but not core protein, modulates the efficiency of cap-independent translation directed by the internal ribosome entry site of hepatitis C virus. J Virol 74:11347–11358
Lytle JR, Wu L, Robertson HD (2002) Domains on the hepatitis C virus internal ribosome entry site for 40S subunit binding. RNA 8:1045–1055
Ji H, Fraser CS, Yu Y, Leary J, Doudna JA (2004) Coordinated assembly of human translation initiation complexes by the hepatitis C virus internal ribosome entry site RNA. Proc Natl Acad Sci USA 101:16990–16995
Otto GA, Puglisi JD (2004) The pathway of HCV IRES-mediated translation initiation. Cell 119:369–380
Ito T, Tahara SM, Lai MM (1998) The 3’-untranslated region of hepatitis C virus RNA enhances translation from an internal ribosomal entry site. J Virol 72:8789–8796
Bradrick SS, Walters RW, Gromeier M (2006) The hepatitis C virus 3’-untranslated region or a poly(A) tract promote efficient translation subsequent to the initiation phase. Nucleic Acids Res 34:1293–1303
Morikawa K, Ito T, Nozawa H, Inokuchi M, Uchikoshi M, Saito T, Mitamura K, Imawari M (2006) Translational enhancement of HCV RNA genotype 1b by 3’-untranslated and envelope 2 protein-coding sequences. Virology 345:404–415
Lourenço S, Costa F, Debarges B, Andrieu T, Cahour A (2008) Hepatitis C virus internal ribosome entry site-mediated translation is stimulated by cis-acting RNA elements and trans-acting viral factors. FEBS J 275:4179–4197
Bung C, Bochkaeva Z, Terenin I, Zinovkin R, Shatsky IN, Niepmann M (2010) Influence of the hepatitis C virus 3’-untranslated region on IRES-dependent and cap-dependent translation initiation. FEBS Lett 584:837–842
Petrik J, Parker H, Alexander GJ (1999) Human hepatic glyceraldehyde-3-phosphate dehydrogenase binds to the poly(U) tract of the 3’ non-coding region of hepatitis C virus genomic RNA. J Gen Virol 80:3109–3113
Spangberg K, Goobar-Larsson L, Wahren-Herlenius M, Schwartz S (1999) The La protein from human liver cells interacts specifically with the U-rich region in the hepatitis C virus 3’ untranslated region. J Hum Virol 2:296–307
Wood J, Frederickson RM, Fields S, Patel AH (2001) Hepatitis C virus 3’X region interacts with human ribosomal proteins. J Virol 75:1348–1358
McCaffrey AP, Ohashi K, Meuse L, Shen S, Lancaster AM, Lukavsky PJ, Sarnow P, Kay MA (2002) Determinants of hepatitis C translational initiation in vitro, in cultured cells and mice. Mol Ther 5:676–684
Huang L, Hwang J, Sharma SD, Hargittai MR, Chen Y, Arnold JJ, Raney KD, Cameron CE (2005) Hepatitis C virus non-structural protein 5A (NS5A) is a RNA-binding protein. J Biol Chem 280:36417–36428
Wang H, Shen XT, Ye R, Lan SY, Xiang L, Yuan ZH (2005) Roles of the polypyrimidine tract and 3’ noncoding region of hepatitis C virus RNA in the internal ribosome entry site-mediated translation. Arch Virol 150:1085–1099
Scheller N, Mina LB, Galao RP, Chari A, Gimenez-Barcons M, Noueiry A, Fischer U, Meyerhans A, Díez J (2009) Translation and replication of hepatitis C virus genomic RNA depends on ancient cellular proteins that control mRNA fates. Proc Natl Acad Sci USA 106:13517–13522
Weinlich S, Huttelmaier S, Schierhorn A, Behrens SE, Ostareck-Lederer A, Ostareck DH (2009) IGF2BP1 enhances HCV IRES-mediated translation initiation via the 3’UTR. RNA 15:1528–1542
Yu KL, Jang SI, You JC (2009) Identification of in vivo interaction between Hepatitis C Virus core protein and 5’ and 3’ UTR RNA. Virus Res 145:285–292
Spangberg K, Wiklund L, Schwartz S (2001) Binding of the La autoantigen to the hepatitis C virus 3’ untranslated region protects the RNA from rapid degradation in vitro. J Gen Virol 82:113–120
Sachs AB, Sarnow P, Hentze MW (1997) Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell 89:831–838
Edgil D, Harris E (2006) End-to-end communication in the modulation of translation by mammalian RNA viruses. Virus Res 119:43–51
Harris E, Holden KL, Edgil D, Polacek C, Clyde K (2006) Molecular biology of flaviviruses. Novartis Found Symp 277:23–39 Discussion 40, 71–23, 251–253
Serrano P, Pulido MR, Saiz M, Martínez-Salas E (2006) The 3’ end of the foot-and-mouth disease virus genome establishes two distinct long-range RNA–RNA interactions with the 5’ end region. J Gen Virol 87:3013–3022
Romero-López C, Berzal-Herranz A (2009) A long-range RNA–RNA interaction between the 5’ and 3’ ends of the HCV genome. RNA 15:1740–1752
Kolupaeva VG, Pestova TV, Hellen CU (2000) An enzymatic footprinting analysis of the interaction of 40S ribosomal subunits with the internal ribosomal entry site of hepatitis C virus. J Virol 74:6242–6250
Lukavsky PJ, Otto GA, Lancaster AM, Sarnow P, Puglisi JD (2000) Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function. Nat Struct Biol 7:1105–1110
Babaylova E, Graifer D, Malygin A, Stahl J, Shatsky I, Karpova G (2009) Positioning of subdomain IIId and apical loop of domain II of the hepatitis C IRES on the human 40S ribosome. Nucleic Acids Res 37:1141–1151
Barria MI, González A, Vera-Otarola J, Leon U, Vollrath V, Marsac D, Monasterio O, Pérez-Acle T, Soza A, López-Lastra M (2009) Analysis of natural variants of the hepatitis C virus internal ribosome entry site reveals that primary sequence plays a key role on cap-independent translation. Nucleic Acids Res 37:957–971
Jubin R, Vantuno NE, Kieft JS, Murray MG, Doudna JA, Lau JY, Baroudy BM (2000) Hepatitis C virus internal ribosome entry site (IRES) stem loop IIId contains a phylogenetically conserved GGG triplet essential for translation and IRES folding. J Virol 74:10430–10437
Klinck R, Westhof E, Walker S, Afshar M, Collier A, Aboul-Ela F (2000) A potential RNA drug target in the hepatitis C virus internal ribosomal entry site. RNA 6:1423–1431
Lee H, Shin H, Wimmer E, Paul AV (2004) cis-acting RNA signals in the NS5B C-terminal coding sequence of the hepatitis C virus genome. J Virol 78:10865–10877
You S, Stump DD, Branch AD, Rice CM (2004) A cis-acting replication element in the sequence encoding the NS5B RNA-dependent RNA polymerase is required for hepatitis C virus RNA replication. J Virol 78:1352–1366
Friebe P, Boudet J, Simorre JP, Bartenschlager R (2005) Kissing-loop interaction in the 3’ end of the hepatitis C virus genome essential for RNA replication. J Virol 79:380–392
Romero-López C, Díaz-González R, Berzal-Herranz A (2007) Inhibition of hepatitis C virus internal ribosome entry site-mediated translation by an RNA targeting the conserved IIIf domain. Cell Mol Life Sci 64:2994–3006
Romero-López C, Barroso-delJesus A, Puerta-Fernández E, Berzal-Herranz A (2005) Interfering with hepatitis C virus IRES activity using RNA molecules identified by a novel in vitro selection method. Biol Chem 386:183–190
Romero-López C, Díaz-González R, Barroso-delJesus A, Berzal-Herranz A (2009) Inhibition of HCV replication and IRES-dependent translation by an RNA molecule. J Gen Virol 90:1659–1669
Diviney S, Tuplin A, Struthers M, Armstrong V, Elliott RM, Simmonds P, Evans DJ (2008) A hepatitis C virus cis-acting replication element forms a long-range RNA–RNA interaction with upstream RNA sequences in NS5B. J Virol 82:9008–9022
Komarova AV, Brocard M, Kean KM (2006) The case for mRNA 5’ and 3’ end cross talk during translation in a eukaryotic cell. Prog Nucleic Acid Res Mol Biol 81:331–367
Corver J, Lenches E, Smith K, Robison RA, Sando T, Strauss EG, Strauss JH (2003) Fine mapping of a cis-acting sequence element in yellow fever virus RNA that is required for RNA replication and cyclization. J Virol 77:2265–2270
Markoff L (2003) 5’- and 3’-noncoding regions in flavivirus RNA. Adv Virus Res 59:177–228
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415
Junemann C, Song Y, Bassili G, Goergen D, Henke J, Niepmann M (2007) Picornavirus internal ribosome entry site elements can stimulate translation of upstream genes. J Biol Chem 282:132–141
Henke JI, Goergen D, Zheng J, Song Y, Schuttler CG, Fehr C, Junemann C, Niepmann M (2008) microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J 27:3300–3310
Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P (2005) Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science 309:1577–1581
Cai Z, Zhang C, Chang KS, Jiang J, Ahn BC, Wakita T, Liang TJ, Luo G (2005) Robust production of infectious hepatitis C virus (HCV) from stably HCV cDNA-transfected human hepatoma cells. J Virol 79:13963–13973
Wakita T, Pietschmann T, Kato T et al (2005) Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med 11:791–796
Boonstra A, van der Laan LJ, Vanwolleghem T, Janssen HL (2009) Experimental models for hepatitis C viral infection. Hepatology 50:1646–1655
Acknowledgments
The Huh-7 cell line was a kind gift of Dr. R. Aldabe. We thank Dr. Alicia Barroso-delJesus for helpful discussions. We also thank Vicente Augustin for excellent technical assistance. This work was supported by Grants BFU2009-08137 from the Spanish Ministerio de Innovación y Ciencia, CTS-5077 from the Junta de Andalucía and FEDER funds from the EU to A.B–H. C.R-L was funded by Grants 2004-20E632 from the Spanish National Research Council (CSIC) and BFU2009-08137.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Romero-López, C., Berzal-Herranz, A. The functional RNA domain 5BSL3.2 within the NS5B coding sequence influences hepatitis C virus IRES-mediated translation. Cell. Mol. Life Sci. 69, 103–113 (2012). https://doi.org/10.1007/s00018-011-0729-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-011-0729-z