Summary
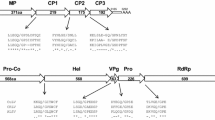
The 3699 nt genome of olive latent virus 1 (OLV-1), described years ago from Southern Italy as a putative sobemovirus, was completely sequenced. OLV-1 genomic RNA was not polyadenylated and had a structure virtually identical to that of species of theNecrovirus rather than theSobemovirus genus. Five open reading frames (ORFs) were identified, of which the 5′-proximal encoded a 23 K protein and ended with an amber codon whose readthrough could yield a putative 82 K product. This polypeptide had extensive sequence similarity with polymerases of serotypes A and D of tobacco necrosis necrovirus (TNV-A and TNV-D) and species of the familyTombusviridae and related genera (Dianthovirus andMachlomovirus). Two small ORFs followed, which encoded polypeptides of 8 K and 6 K, respectively. The 6 K product had extensive homology with the comparable 6 K protein of TNV-A and was also related to the 11 K protein of shallot latent carlavirus, one of the “triple block” polypeptides involved in cell-to-cell virus movement. The 3′-proximal ORF was in the same position as the coat protein (CP) cistron of necroviruses and encoded a 30 K product related to CP of both TNV-A and -D. Computer-assisted comparative analysis of structural and non-structural proteins of OLV-1, TNV-A and TNV-D disclosed an overall distant relationship between OLV-1 and TNV-D. OLV-1 genome appeared homologous to that of TNV-A, but differences from TNV-A were the absence of the small ORF downstream of the CP cistron and in the low degree of sequence identity in CP (39% aa identity). OLV-1 is serologically distantly related to TNV-A and even more distantly related to TNV-D. We propose that OLV-1 is a necrovirus species in its own right.
Similar content being viewed by others
References
Agranovsky AA, Koenig R, Maiss E, Boyko VP, Casper R, Atabekov JG (1994) Expression of beet yellows closterovirus capsid protein and p24, a capsid protein homologue in vitro and in vivo. J Gen Virol 75: 1431–1439
Argos P (1988) A sequence motif in many polymerases. Nucleic Acids Res 16: 9909–9916
Beck DL, Guilford PJ, Voot DM, Andersen MT, Forster RSL (1991) Triple gene block proteins of white clover mosaic potexvirus are required for transport. Virology 183: 695–702
Carrington JC, Heaton LA, Zuidema D, Hillman BI, Morris TJ (1989) The genome structure of turnip crinkle virus. Virology 170: 219–226
Coutts RHA, Ridgen JE, Slabas AR, Lomonossoff GP, Wise PJ (1991) The complete nucleotide sequence of tobacco necrosis virus strain D. J Gen Virol 72: 1521–1529
Çinar A, Kersting U, Önelge N, Korkmaz S, Sas G (1993) Citrus virus and virus-like diseases in the eastern Mediterranean region of Turkey. Proc 12th IOCV Conference, Riverside, pp 397–400
Felsenstein J (1989) PHYLIP-phylogeny inference package (version 3.5). Cladistics 5: 164–166
Frohman MA, Dush MK, Martin GR (1988) Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA 85: 8998–9002
Gallie DR, Sleat DE, Watts JW, Turner PC, Wilson TMA (1987) A comparison of eucariotic viral 5′-leader sequences as enhancers of mRNA expression in vivo. Nucleic Acids Res 17: 8693–8711
Gallitelli D, Savino V (1985) Olive latent virus 1, an isometric virus with a single RNA species isolated from olive in Apulia, Southern Italy. Ann Appl Biol 106: 295–303
Goldbach R, Le Gall O, Wellink J (1991) Alpha-like viruses in plants. Semin Virol 2: 19–25
Grieco F, Burgyan J, Russo M (1989) Nucleotide sequence of the 3′-terminal region of cymbidium ringspot virus RNA. J Gen Virol 70: 2533–2538
Grieco F, Burgyan J, Russo M (1989) The nucleotide sequence of cymbidium ringspot virus RNA. Nucleic Acids Res 17: 6383
Grieco F, Cillo F, Barbarossa L, Gallitelli D (1992) Nucleotide sequence of a cucumber mosaic virus satellite RNA, associated with a tomato top stunting. Nucleic Acids Res 20: 6733
Grieco F, Martelli GP, Savino V (1995) The nucleotide sequence of RNA3 and RNA4 of olive latent virus 2. J Gen Virol 76: 929–937
Guilley H, Carrington C, Balazs E, Jonard G, Richards K, Morris TJ (1985) Nucleotide sequence and genome organization of carnation mottle virus RNA. Nucleic Acids Res 13: 6663–6677
Hattori M, Sakaki Y (1986) Dideoxy sequencing method using denatured plasmid templates. Anal Biochem 152: 232–238
Higgins DG, Sharp PM (1988)Clustal: a package for performing multiple sequence alignment on a microcomputer. Gene 73: 237–244
Hull R (1995) GenusSobemovirus In: Murphy FA, Fauquet CM, Bishop DHL, Ghabrial SA, Jarvis AW, Martelli GP, Mayo MA, Summers MD (eds) Virus Taxonomy. Classification and Nomenclature of Viruses. Sixth Report of International Committee on Taxonomy of Viruses. Springer, Wien New York, pp 376–378 (Arch Virol [Suppl] 10)
Jayarama Bhat G, Lodes MJ, Myler PJ, Stuart KD (1991) A simple method for cloning blunt ended DNA fragments. Nucleic Acids Res 19: 398
Kanyuka KV, Vishnichenko VK, Levay KE, Kondrikov DY, Ryabov EV, Zavriev SK (1992) Nucleotide sequence of shallot virus X RNA reveals 5′-proximal cistron closely related to those of potyviruses and a unique arrangement of the 3′ proximal cistrons. J Gen Virol 73: 2553–2560
Koonin E (1991) The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J Gen Virol 72: 2197–2206
Koonin E, Dolja VV (1993) Evolution and taxonomy of positive-strand viruses: implications of comparative amino acid sequences. Crit Rev Biochem Mol Biol 28: 375–430
Lutcke HA, Chow KC, Mickel FS, Moss KA, Kern HF, Scheele GA (1987) Selection of AUG initiation codons differs in plant and animals. EMBO J 6: 43–48
Marck C (1988) “DNA Strider”: a “C” programme for the fast analysis of DNA and protein sequences on the Apple Macintosh family computers. Nucleic Acids Res 16: 1829–1836
Martelli GP (1992) Classification and nomenclature of plant viruses: state of the art. Plant Dis 76: 436–442
Martelli GP, Russo M (1995) FamilyTombusviridae In: Murphy FA, Fauquet CM, Bishop DHL, Ghabrial SA, Jarvis AW, Martelli GP, Mayo MA, Summers MD (eds) Virus Taxonomy. Classification and Nomenclature of Viruses. Sixth Report of International Committee on Taxonomy of Viruses. Springer, Wien New York, pp 392–397 (Arch Virol [Suppl] 10)
Martelli GP, Sabanadzovic S, Savino V, Abu-Zurayk AR, Masannat K (1995) Virus-like diseases and viruses of olive in Jordan. Phytopathol Medit 34: 133–136
Martelli GP, Yilmaz MA, Savino V, Baloglou S, Grieco F, Güldür MA, Boscia D, Greco N, Lafortezza R (1995) Properties of a citrus isolate of olive latent virus 1, a seemingly new necrovirus. Eur J Plant Pathol (in press)
Meulewaeter F, Seurinck J, Van Emmelo J (1990) Genome structure of tobacco necrosis virus A. Virology 117: 699–709
Meulewaeter F, Cornelissen M, Van Emmelo J (1994) Suggenomic RNAs mediated expression of cistons located internally on the genomic RNA of tobacco necrosis virus strain A. J Virol 66: 6419–6428
Meulewaeter F, Danthinne X, Van Emmelo J (1994) Necroviruses. In: Webster RG, Granoff A (eds) Encyclopedia of virology, vol 2. Academic Press, New York, 896–901
Morris TJ, Dodds JA (1979) Isolation and analysis of double-stranded RNA from virus-infected plant and fungal tissue. Phytopathology 69: 854–858
Nicolas O, Lalibertè JF (1992) The complete nucleotide sequence of turnip mosaic potyvirus RNA. J Gen Virol 73: 2785–2793
Pearson WR, Lipman DJ (1988) Improved tools for biological sequence comparison. Proc Natl Acad Sci USA 85: 2444–2448
Program Manual for the Wisconsin Package, Version 8, September 1994, Genetics Computer Group, 575 Science Drive, Madison, Wisconsin, USA 53711
Riviere CJ, Rochon DM (1990) Nucleotide sequence and genomic organization of melon necrotic spot virus. J Gen Virol 72: 1887–1896
Russo M, Burgyan J, Martelli GP (1994) Molecular biology ofTombusviridae. Adv Virus Res 44: 381–428
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York
Skotnicki ML, Mackenzie AM, Torronen M, Gibbs AJ (1993) The genomic sequence of cardamine chlorotic fleck carmovirus. J Gen Virol 74: 1933–1937
Smith TF, Waterman MS (1981) Identification of common molecular subsequences. J Mol Biol 147: 195–197
White JL, Kaper JM (1989) A simple method for detection of viral satellite RNAs in small plant tissue samples. J Virol Methods 23: 83–94
Wu S, Rinehart CA, Kaesberg P (1987) Sequence and genome organization of southern bean mosaic virus genomic RNA. Virology 161: 73–80
Zhang L, French R, Langenberg WG (1993) Molecular cloning and sequencing of the coat protein gene of a Nebraskan isolate of tobacco necrosis virus: the deduced coat protein sequence has only moderate homology with those of strain A and strain D. Arch Virol 132: 291–305
Author information
Authors and Affiliations
Additional information
The sequence data reported in this paper have been deposited in the EMBL database and given the accession number X85989.
Rights and permissions
About this article
Cite this article
Grieco, F., Savino, V. & Martelli, G.P. Nucleotide sequence of the genome of a citrus isolate of olive latent virus 1. Archives of Virology 141, 825–838 (1996). https://doi.org/10.1007/BF01718158
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF01718158