Abstract
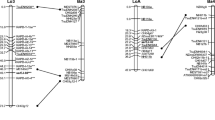
We have established the first linkage map forPetunia hybrida based upon both RAPD and phenotypical markers. The progeny studied consisted of 100 BC1 individuals derived from the [(St40xTlvl)xTlvl] back-cross. Each morphological marker has previously been mapped onto one of the seven chromosomes. The map consists of 35 RAPD loci of which 24 were affected onto chromosomes while 10 loci were not affected. The loci covered 262.9 cM with a mean distance of 8.2 cM. They are dispersed over seven linkage groups, of which six are carried on identified chromosomes. The RAPD markers were also applied on a set of tenP. hybrida, lines chosen for their diversity and on a set of seven wild species corresponding to the possible ancestors of theP. hybrida species. The markers were found both in the wild species as well as inP. hybrida lines indicating that they are inherited and are stable enough to establish similarities and to suggest relationships between species. Eight out of the ten lines carry different linkage groups of RAPD markers, which suggest that recombinant events occurred between chromosomes which originated in the wild species.
Similar content being viewed by others
References
Bernatsky R, Tanksley SD (1986) Toward a saturated linkage map in tomato based on isozymes and random cDNA sequences. Genetics 112:887–898
Bianchi F, Dommergues P (1979)Petunia genetics.Petunia as a model for plant research genetics and mutagenesis. Ann Amélior Plant 29:607–610
Cornu A (1984) Genetics ofPetunia. In: Monographs on theoretical and applied genetics 9. Sink KC (ed), Springer, Berlin Heidelberg New York Tokyo, pp 34–48
Cornu A, Maizonnier D, Wiering H, de Vlaming P (1980) Petunia genetics. III. The linkage group of chromosome V. Ann Amélior Plant 30:443–454
Cornu A, Farcy E, Maizonnier D, Haring M, Veerman W, Geratz AGM (1990)Petunia hybrida. In: O'Brien SJ (ed) Genetic maps. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York, pp 107–124
Echt CS, Erdahl A, Mc Coy TJ (1992) Genetic segregation of random amplified polymorphic DNA in diploid cultivated alfalfa. Genome 35:84–87
Folkerts O, Hanson MR (1989) Three copies of a single recombination repeat occur on the 443-kb mastercircle of thePetunia hybrida 3704 mitochondrial genome. Nucleic Acids Res 18: 7345–7357
Hanson MR (1980)Petunia as a model system for molecular biologists. Plant Mol Biol Newslett 1:1–37
Havey MJ, Muehlbauer FJ (1989) Linkages between restriction fragment length polymorphisms, isozyme, and morphological markers in lentil. Theor Appl Genet 77:395–401
Klein-Lankhorst RM, Vermunt A, Weide R, Liharska T, Zabel P (1991) Isolation of molecular markers for tomato (L. esculentum) using random amplified polymorphic DNA (RAPD). Theor Appl Genet 83:108–114
Koes RE, Spelt CE, Mol JNM, Geratz AGM (1987) The chalconesynthase multigene family ofPetunia hybrida: sequence homology, chromosomal location and evolutionary aspects. Plant Mol Biol 10:375–385
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln S and Newburg L (1987) MAPMAKER: An interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Landry BS, Kesseli RV, Farrara B, Michelmore RW (1987) A genetic map of lettuce (Lactuca sativa L.) with restriction fragment length polymorphism, isozyme, disease resistance and morphological markers. Genetics 116:331–337
Lincoln S.E, MJ Daly and ES Lander (1990) MAPMAKER 2.0 Reference manual. Whitehead Institute for Biomedical Research technical report
Maizonnier D (1976) Production de tétraploïdes et de trisomiques naturels chez le Pétunia. Ann Amélior Plant 26:305–318
Maizonnier D (1984). Cytology, chapter 4 In: Sink KC (ed.) Petunia. Monographs on theoretical and applied genetics 9. Springer, Berlin Heidelberg New York Tokyo, pp 21–34
Maizonnier D, Cornu A (1971) A telocentric translocation responsible for variegation in Petunia. Genetica 42:422–436
Maizonnier D, Cornu A (1979) Preuve cytogénétique de la production de chromosomes linéaires remaniés à partir d'un chromosome annulaire chezPetunia hybrida Hort. Caryologia 32:393–412
Maizonnier D, Moessner A (1979) Localization of the seven linkage groups inPetunia hybrida. Genetica 51:143–148
Maizonnier D, Cornu A, Farcy E, De Vlaming P (1986) Genetic and cytological maps in Petunia. In: Horn W, Jensen CJ, Odenbach W, Schieder O (eds). Genetic manipulation in plant breeding. Walter de Gruyter, Berlin NewYork, pp 183–186
McLean M, Geratz AGM, Baird WV, Meagher RB (1990) Six actin gene subfamilies map to five chromosomes ofPetunia hybrida. J Hered 81:341–346
Palmer JD (1983) Chloroplast DNA evolution and biosystematic uses of chloroplast DNA variation. Am Nat 130: S6-S29
Penner GA, Bush A, Wise R, Kim W, Domier L, Kasha K, Laroche A, Scoles G, Molnar SJ, Fedak G (1993) Reproducibility of random amplified polymorphic DNA (RAPD) analysis among laboratories. PCR Meth Appl 2:341–345
Reiter RS, Williams JGK, Feldmann KA, Rafalski JA (1992) Global and local genome mapping inArabidopsis thaliana by using recombinant inbred and random amplified polymorphic DNAs. Proc Natl Acad Sci USA 89:1477–1481
Sederoff R, Neale D (1991) Genome mapping in pines takes shape. Probe vol 1 no 3/4:1–3
Sink KC (1984). Taxonomy chap.2 in: Petunia. Monographs on theoretical and applied genetics 9. Sink KC (ed), Springer, Berlin Heidelberg New York Tokyo, pp 3–7
Stout AB (1952) Reproduction inPetunia. Mem Torrey Bot Club 20:1–202
Systma KJ, Gottlieb LD (1986) Chloroplast DNA evolution and phylogenetic relationships inClarkia Sect.peripetasma (Onagraceae). Evolution 40:1248–1261
Tautz D, Renz M (1983) An optimized squeeze-freeze method for the recovery of DNA from agarose gels. Anal Biochem 132:14–19
Vierling RA, Nguyen HT (1992) Use of RAPD markers to determine the genetic diversity of diploid, wheat genotypes. Theor Appl Genet 84:835–838
Wiering H. De Vlaming P, Cornu A, Maizonnier D (1979)Petunia genetics. I. List of genes. Ann Amélior Plant 29:611–622
Wijsman (1982) On the inter-relationships of certain species ofPetunia. I. Taxonomic notes on the parental species ofPetunia hybrida. Acta Bot Neerl 31:477–490
Williams JGK, Kubelic AR, Livak KJ, Rafalski JA, Tingey SV (1990) DNA polymorphisms amplified by arbitrary primers are useful as gentic markers. Nucleic Acids Res 18:6531–6535
Author information
Authors and Affiliations
Additional information
Communicated by H. F. Linskens
Rights and permissions
About this article
Cite this article
Peltier, D., Farcy, E., Dulieu, H. et al. Origin, distribution and mapping of RAPD markers from wildPetunia species inPetunia hybrida Hort lines. Theoret. Appl. Genetics 88, 637–645 (1994). https://doi.org/10.1007/BF01253965
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF01253965