Abstract
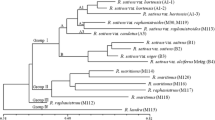
DNA polymorphisms among Arabidopsis thaliana ecotypes are widely used as genetic markers in map-based cloning strategies. New PCR-based molecular markers do not only facilitate molecular mapping, but can also be used to obtain reliable sequence information for cladistic analyses. We have used CAPS (cleaved amplified polymorphic sequences) markers and a direct sequencing strategy to estimate genetic similarity among eighteen Arabidopsis ecotypes. Sequences at four loci, two from the nuclear and two from a non-nuclaar genome, were analysed. For each ecotype more than 1000pb of sequence information was obtained, and genetic similarity was calculated from a total of 35 polymorphic sites using a character-based approach. Divergence ranged from zero up to 50 discordant characters among the 72 characters defined by the polymorphisms. Separate calculations based on the nuclear and the non-nuclear sequences were performed and revealed a number of common features, including the existence of small clusters of very closely related ecotypes separated from each other by extensive sequence divergence. Our results provide information useful especially to investigators setting up crosses for chromosome landing strategies.
Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ: Basic local alignment search tool. J Mol Biol 215: 403–410 (1990).
Chang C, Bowman JL, DeJohn AW, Lander ES, Meyerowitz EM: Restriction fragment length polymorphism linkage map for Arabidopsis thaliana. Proc Natl Acad Sci 85: 6856–6860 (1988).
Clegg MT, Gaut BS, Mearn GHJr, Morton BR: Rates and patterns of chloroplast DNA evolution. Proc Natl Acad Sci USA 91: 6795–6801 (1994).
Curtis SE, Clegg MT: Molecular evolution of chloroplast DNA sequences. Mol Biol Evol 1: 291–301 (1984).
Ellsworth DL, Rittenhouse KD, Honeycutt RL: Artifactual variation in randomly amplified polymorphic DNA banding patterns. Biotechniques 14: 214–217 (1993).
Gonzalez-Candelas F, Elena SF, Moya A: Approximate variance of nucleotide divergence between two sequences estimated from restriction fragment data. Genetics 140: 1443–1446 (1995).
Grill E, Somerville C: Construction and characterization of a yeast artificial chromosome library of Arabidopsis thaliana which is suitable for chromosome walking. Mol Gen Genet 226: 484–490 (1991).
Grover NS: Characterization of Arabidopsis thaliana ecotypes on the basis of genetic variation at ten isozyme loci. Arabidopsis Inf Serv 12: 19–21 (1975).
Hanfstingl U, Berry A, Kellogg EA, Costa JTIII, Rüdiger W, Ausubel FM: Haplotypic divergence coupled with lack of diversity at the Arabidopsis thaliana alcohol dehydrogenase locus: roles for both balancing and directional selection? Genetics 138: 811–828 (1994).
Hardtke CS, Berleth T: Genetic and contig map of a 2200 kb region encompassing 5.5 cM on chromosome 1 of Arabidopsis thaliana. Genome, in press.
Hillis DM, Allard MW, Miyamoto MM: Analysis of DNA sequence data: phylogenetic inference. Meth Enzymol 224: 456–487 (1990).
King G, Nienhuis J, Hussey C: Genetic similarity among ecotypes of Arabidopsis thaliana estimated by analysis of restriction fragment length polymorphisms. Theor Appl Genet 86: 1028–1032 (1993).
Klein M, Eckert-Ossenkopp U, Schmiedeberg I, Brandt P, Unseld M, Brennicke A, Schuster W: Physical mapping of the mitochondrial genome of Arabidopsis thaliana by cosmid and YAC clones. Plant J 6: 447–455 (1994).
Konieczny A, Ausubel FM: A procedure for mapping Arabidopsis thaliana mutations using co-dominant ecotypespecific PCR-based markers. Plant J 4: 403–410 (1993).
Murray HG, Thompson WF: Rapid isolation of high molecular weight plant DNA. Nucl Acids Res 8: 4321–4325 (1980).
Nam H, Giraudat J, denBoer B, Moonan F, Loos WDB, Hauge BM, Goodman HM: Restriction fragment length polymorphism linkage map of Arabidopsis thaliana. Plant Cell 1: 699–705 (1989).
Pearson WR, Lipman DJ: Improved tools for biological sequence comparison. Proc Natl Acad Sci USA 85: 2444–2448 (1988).
Penner GA, Bush A, Wise R, Kim W, Domier L, Kasha K, Laroche A, Scoles G, Molnar SJ, Fedak G: Reproducibility of random amplified polymorphic DNA (RAPD) analysis among laboratories. PCR Meth Appl 2: 341–345 (1993).
Reiter RS, Williams JGK, Feldmann KA, Rafalski J, Tingey SV, Scolnik P: Global and local genome mapping in Arabidopsis thaliana by using recombinant inbred lines and random amplified polymorphic DNAs. Proc Natl Acad Sci USA 89: 1477–1481 (1992).
Schmidt R, Putterill J, West J, Cnops G, Robson F, Coupland G, Dean C: Analysis of clones carrying repeated DNA sequences in two YAC libraries of Arabidopsis thaliana DNA. Plant J 5: 735–744 (1994).
Swofford DL: Phylogenetic analysis using parsimony, version 3.0, computer program. Illinois Natural History Survey, Champaign, IL (1990).
Tagaki S, Kimura M, Katsuki M: Direct sequencing of PCR products using unlabeled primers. Biotechniques 14: 218–221 (1993).
Thomas WK, Kocher TD: Sequencing of PCR amplified DNAs. Meth Enzymol 224: 391–399 (1993).
Vos P, Hogers R, Bleeker M, Reijans M, van deLee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M: AFLP: a new technique for DNA fingerprinting. Nucl Acids Res 23: 4407–4414 (1995).
Williams JG, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV: DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucl Acids Res 18: 6531–6535 (1990).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Hardtke, C.S., Müller, J. & Berleth, T. Genetic similarity among Arabidopsis thaliana ecotypes estimated by DNA sequence comparison. Plant Mol Biol 32, 915–922 (1996). https://doi.org/10.1007/BF00020488
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00020488