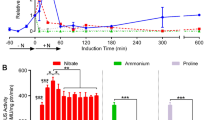
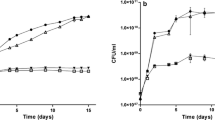
Abstract
Expression ofnit-3 andnit-6, the structural genes which encode nitrate reductase and nitrite reductase inNeurospora crassa, requires the global-acting NIT2 and the pathway specific NIT4 regulatory proteins. NIT4, which consists of 1090 amino-acid residues, possesses a Cys6/Zn2 zinc cluster DNA-binding-domain. NIT4 was dissected to localize transactivation domains by fusion of various segments of NIT4 to the DNA-binding domain of GAL4 for in vivo analysis in yeast. Three separate activation subdomains, and one negative-acting region, which function in yeast were located in the carboxyl-terminal region of NIT4. The C-terminal tail of 28 amino-acid residues was identified as a minimal activation domain and consists of a novel leucine-rich, acidic region. Most deletions which removed even small segments of the NIT4 protein were found to lead to the loss of NIT4 function in vivo inN. crassa, implying that the central region of the protein which lies between the DNA-binding and activation domains is essential for function. The yeast two-hybrid system was employed to identify regions of NIT4 responsible for dimer formation. A short isoleucine-rich segment downstream from the zinc cluster, predicted to form a coiled coil, allowed dimerization in vivo; this same isoleucine-rich region also showed dimerization in vitro when examined via chemical cross linking. The enzyme nitrate reductase has been postulated to exert autogenous regulation by directly interacting with the NIT4 protein. This possible nitrate reductase-NIT4 interaction was investigated with the yeast two-hybrid system and by direct in vitro binding assays; both assays failed to identify such a protein-protein interaction.
Similar content being viewed by others
References
Attardi LD, Tjian R (1993)Drosophila tissue-specific transcription factor NTF-1 contains a novel isoleucine-rich activation motif. Genes Dev 7:1341–1353
Ausubel FM, Brent R, Kingston RE, et al. (1987) Current protocols in molecular biology, John Wiley and Sons, New York
Chien CT, Bartel PL, Sternglanz R, Fields S (1991) The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci USA 88:9578–9582
Cook WJ, Chase E, Audino DC, Dennis CL (1994) Dissection of the ADR1 protein reveals multiple, functionally redundant activation domains interspersed with inhibitory regions: evidence for a repressor binding to the ADR1c region. Mol Cell Biol 14:629–640
Cress WD, Triezenberg SJ (1991) Critical structural elements of the VP16 transcriptional activation domain. Science 251:87–90
Davis RH, deSerres FJ (1970) Genetic and microbiological research techniques forNeuropsora crassa. Methods Enzymol 17A:79–143
Durfee T, Becherer K, Chen PL, Yeh SH, Yang YZ, Kilburn AE, Lee WH, Elledge SJ (1994) The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev 7:555–569
Ferreri K, Gill G, Montminy M (1994) The cAMP-regulated transcription factor CREB interacts with a component of the TFIID complex. Proc Natl Acad Sci USA 91:1210–1213
Fields S, Song OK (1989) A novel genetic system to detect protein-protein interaction. Nature 340:245–246
Fu YH, Marzluf GA (1988) Metabolic control and autogenous regulation of nit-3 the nitrate reductase structural gene ofNeurospora crassa. J Bacteriol 170:657–661
Fu YH, Feng B, Evans S, Marzluf GA (1995) Sequence-specific DNA binding by NIT4, the pathway-specific regulatory protein which mediates nitrate induction in Neurospora. Mol Microbiol 15:935–942
Gerber HP, Seipel K, Georgiev O, Hofferer M, Hug M, Rusconi S, Schaffner W (1994) Transcriptional activation modulated by polymeric glutamine and proline. Science 263:808–810
Gill G, Pascal E, Tseng AH, Tjian R (1994) A glutamine-rich hydrophobic patch in transcription factor SP1 contacts the dTAFII-110 component of theDrosophila TFIID complex and mediates transcriptional activation. Proc Natl Acad Sci USA 91:192–196
Goodrich JA, Hoey T, Thut CJ, Admon A, Tjian R (1993)Drosophila TAFII40 interacts with both a VP16 activation domain and the basal transcription factor TFIIB. Cell 75:519–530
Henikoff S (1984) Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene 28:351–358
Hope IA, Struhl K (1986) Functional dissection of a eukaryotic transcriptional activator protein, GCN4, of yeast. Cell 46:885–894
Hope IA, Mahodevan S, Struhl K (1988) Structural and functional characterization of the short acidic transcriptional activation region of yeast GCN4 protein. Nature 333:635–640
Hoy MV, Leuther KK, Kodadek T, Johnston SA (1993) The acidic activation domains of the GDN4 and GAL4 proteins are not alpha helical but form beta sheets. Cell 72:587–594
Kanaan MN, Marzluf GA (1993) The positive-acting sulfur regulatory protein CYS3 ofNeurospora crassa: nuclear localization, autogenous control, and regions required for transcriptional activation. Mol Gen Genet 239:334–344
Kunkel TA (1985) Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA 82:488–492
Landschulz WH, Johnson PF, Mcknight SL (1989) The DNA-binding domain of the rat liver protein c/EBP is bipartite. Science 243:1681–1688
Leuther KK, Salmeron JM, Johnston SA (1993) Genetic evidence that an activation domain of GAL4 does not require acidity and may form a beta-sheet. Cell 72:575–585
Lupas A, Dyke MV, Stock J (1991) Predicting coiled coils from protein sequences. Science 252:1162–1164
Ma J, Ptashne M (1987) Deletion analysis of GAL4 defines two transcriptional activating segments. Cell 48:847–853
Marzluf GA (1981) Regulation of nitrogen metabolism and gene expression in fungi. Microbiol Rev 45:437–461
Marzluf GA (1993) Regulation of sulfur and nitrogen metabolism in filamentous fungi. Annu Rev Microbiol 47:31–55
Meyer ME, Quirin-Stricker C, Lerouge T, Bocquel MT, Gronemeyer H (1992) A limiting factor mediates the differential activation of promoters by the human progesterone receptor isoforms. J Biol Chem 267:10882–10887
Ohashi Y, Brickman JM, Furman E, Middleton B, Carey M (1994) Modulating the potency of an activator in a yeast in vitro transcription system. Mol Cell Biol 14:2731–2739
Ptashne M (1988) How eukaryotic transcriptional activators work. Nature 335:683–689
Qiu HF, Bubois E, Messenguy F (1991) Dissection of the bifunctional ARGRII protein involved in the regulation of arginine anabolic and catabolic pathways. Mol Cell Biol 11:2169–2179
Reece RJ, Ptashne M (1993) Determinants of binding-site specificity among yeast C6 zinc cluster proteins. Science 261:909–911
Schiestl RH, Gietz RD (1989) High-efficiency transformation of intact yeast cells using single-stranded nucleic acids as carrier. Curr Genet 16:339–346
Smith DB, Johnson KS (1988) Single-step purification of polypeptides expressed inE. coli as fusions with glutathione S-transferase. Gene 67:31–40
Studier FW, Moffatt BA (1986) Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol 189:113–130
Tomsett AB, Garrett RH (1981) Biochemical analysis of mutants defective in nitrate assimilation inNeurospora crassa: evidence for autogenous control by nitrate reductase. Mol Gen Genet 184:183–190
Yuan GF, Marzluf GA (1992a) Transformants ofNeurospora crassa with the nit-4 nitrogen regulatory gene: copy number, growth rate and enzyme activity. Curr Genet 22:205–211
Yuan GF, Marzluf GA (1992b) Molecular characterization of mutations of nit-4, the pathway-specific regulatory gene which controls nitrate assimilation inNeurospora crassa. Mol Microbiol 6:67–73
Yuan GF, Fu YH, Marzluf GA (1991) nit-4, a pathway specific regulatory gene ofNeurospora crassa, encodes a protein with a putative binculear zinc DNA-binding domain. Mol Cell Biol 11:5735–5745
Author information
Authors and Affiliations
Additional information
Communicated by K. Esser
Rights and permissions
About this article
Cite this article
Feng, B., Marzluf, G.A. The regulatory protein NIT4 that mediates nitrate induction inNeurospora crassa contains a complex tripartite activation domain with a novel leucine-rich, acidic motif. Curr Genet 29, 537–548 (1996). https://doi.org/10.1007/BF02426958
Received:
Issue Date:
DOI: https://doi.org/10.1007/BF02426958