Abstract
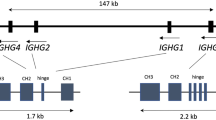
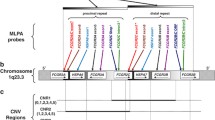
The human immunoglobulin heavy chain constant region locus (IGHC) comprises nine genes and two pseudogenes clustered in a 350 kilobase (kb) region on chromosome 14q32. Several IGHC haplotypes with single or multiple gene deletions and duplications have been characterized. The most likely mechanism accounting for these unusual haplotypes is the unequal crossing-over between homologous regions within the locus. Here we report the analysis of an unusual case of familial clustering of deletions/duplications. In the two branches of the BON family, three duplicated and two deleted haplotypes, all probably independent in origin, have been characterized. The structure of the haplotypes, one of which is described here for the first time, supports the hypothesis of homologous unequal crossing-over as the origin of recombinant haplotypes. The analysis of serological markers in a subject carrying one deleted and one duplicated haplotype allowed us the first direct inferences concerning the functions of the duplicated IGHC haplotypes.
Similar content being viewed by others
References
Bottaro, A., DeMarchi, M., DeLange, G. G., Boccazzi, C., Caldesi, F., Gallina, R., and Carbonara, A. O. New types of multiple and single gene deletions in the human IgCH locus. Immunogenetics 29: 44–48, 1989a
Bottaro, A., DeMarchi, M., DeLange, G. G., and Carbonara, A. O. Gene deletions in human immunoglobulin heavy chain constant region gene cluster. Exp Clin Immunogenet 6: 55–59, 1989b
Bottaro, A., Gallina, R., DeMarchi, M., and Carbonara, A. O. Genetic analysis of new restriction fragment length polymorphisms (RFLP) in the human IgH constant gene locus. Eur J Immunol 19: 2151–2157, 1989c
Bottaro, A., Cariota, U., DeMarchi, M., and Carbonara, A. O. Pulsed field electrophoresis screening for immunoglobulin heavy chain constant region (IGHC) multigene deletions and duplications. Am J Hum Genet 48: 745–756, 1991
Callard, R. E. and Turner, M. W. Cytokines and Ig switching: evolutionary divergence between mice and humans. Immunol Today 11: 200–203, 1990
Carbonara, A. O., Migone, N., Oliviero, S., and DeMarchi, M. Recent advances in primary and acquired immunodeficiences. In F. Aiuti, F. Rosen and M. D. Cooper (eds.): Serono Symposia, vol 28, pp. 219–225, Raven Press, New York, 1986
Engström, P. E., Norhagen, G., Bottaro, A., Carbonara, A. O., Lefranc, G., Steinitz, M., Soder, P. O., Smith, C. I., and Hammarström, L. Subclass distribution of antigen-specific IgA antibodies in normal donors and individuals with homozygous Cα1 or Cα2 gene deletions. J Immunol 145: 109–116, 1990
Flanagan, J. G. and Rabbitts, T. H. Arrangement of human immunoglobulin heavy chain constant region genes implies evolutionary duplication of a segment containing G, E and A genes. Nature 300: 709–713, 1982
Hammarström, L., Carbonara, A. O., DeMarchi, M., Lefranc, G., Lefranc, M. P., and Smith, C. I. E. Generation of the antibody repertoire in individuals with multiple immunoglobulin heavy chain constant region gene deletions. Scand J Immunol 25: 189–194, 1987
Hendriks, R. W., van Tol, M., DeLange, G. G., and Schuurman, R. K. B. Inheritance of a large deletion within the human immunoglobulin heavy chain constant region gene complex and immunological implications. Scand J Immunol 29: 535–541, 1989
Hisajima, H., Nishida, Y., Nakai, S., Takahashi, N., Ueda, S., and Honjo, T. Structure of the human immunoglobulin C E2 pseudogene: implications for its evolutionary origin. Proc Natl Acad Sci USA 80: 2995–2999, 1983
Hofker, M. H., Walter, M. A., and Cox, D. W. Complete physical map of the human immunoglobulin heavy chain constant region gene complex. Proc Natl Acad Sci USA 86: 5567–5571, 1989
Hofker, M. H., Smith, S., Nakamura, Y., Teshima, I., White, R., and Cox, D. W. Physical mapping of probes within 14q32, a subtelomeric region showing a high recombinant frequency. Genomics 6: 33–38, 1990
Kawamura, S., Omoto, K., and Ueda, S. Evolutionary hypervariability in the hinge region of the immunoglobulin alfa gene. J Mol Biol 214: 1–6, 1990
Keyeux, G., Lefranc, G., and Lefranc, M. P. A multigene deletion in the human IGH locus involves highly homologous hot spots of recombination. Genomics 5: 431–441, 1989
Kirsch, I. R., Morton, C. C., Nakahara, K., and Leder, P. Human immunoglobulin heavy chain genes map to a region of translocations in malignant B lymphocytes. Science 216: 301–303, 1982
Lefranc, M. P. and Rabbitts, T. H. Human Immunoglobulin heavy chain A2 gene allotype determination by restriction fragment length polymorphism. Nucleic Acids Res 12: 1303–1311, 1984
Olsson, G., Hofker, M. H., Walter, M. A., Smith, S., Hammarström, L., Smith, C. I. E., and Cox, D. W. Ig H chain variable and C region genes in common variable immunodeficiency. J Immunol 147: 2540–2546, 1991
Rautonen, N., Sarvas, H., and Mäkelä, O. One allele [G2m(n)] of the human IgG2 locus is more productive than the other. Eur J Immunol 19: 817–822, 1989a
Rautonen, N., Seppela, I., Hallberg, T., Grubb, R., and Mäkelä, O. Determination of homozygosity or heterozygosity for the G2m(n) allotype by a monoclonal, precipitating antibody. Exp Clin Immunogenet 6: 31–38, 1989b
Ravetch, J. V., Siebenlist, U., Korsmeyer, S., Waldman, T., and Leder, P. Structure of the human immunoglobulin M locus: characterization of embryonic and rearranged D and J genes. Cell 27: 583–591, 1981
Sambrook, J., Fritsch, E. F., Maniatis, T. Molecular Cloning: A Laborator Manual, 2nd edit, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, USA, 1989
Smith, C. I. E., Hammerström, L., Henter, J. I., and DeLange, G. Molecular and serological analysis of IgG1 deficiency caused by new forms of the constant region of the IGH chain gene deletions. J Immunol 142: 4514–4519, 1989
Takahashi, N., Ueda, S., Obata, M., Nikaido, T., Nakai, S., and Honjo, T. Structure of human immunoglobulin gamma genes: implications for the evolution of a gene family. Cell 29: 671–679, 1982
Ueda, S., Watanabe, Y., Hayashida, H., Miyata, T., Matsuda, F., and Honjo, T. Hominoid evolution based on the structures of immunoglobulin epsilon and alfa genes. Cold Spring Harbor Symposia on Quantitative Biology 51: Cold Spring Harbor Laboratory Press, Cold Spring Harbor, 429–432, 1986
Van Longhem, E. Handbook of Experimental Immunology. In M. Weir (eds.), 3rd edit. Blackwell, Oxford, 1978
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Bottaro, A., Gallina, R., Brusco, A. et al. Familial clustering of IGHC deletions and duplications: functional and molecular analysis. Immunogenetics 37, 356–363 (1993). https://doi.org/10.1007/BF00216800
Received:
Revised:
Issue Date:
DOI: https://doi.org/10.1007/BF00216800