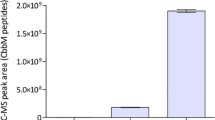
Summary
The cytochrome P450alk gene (P450alk) from Candida tropicalis ATCC 750 was expressed in Saccharomyces cerevisiae GRF18 under control of the alcohol dehydrogenase I (ADHI) promoter. To achieve stable expression over long time periods, a 2-λm derived replicative and an integrative expression system were tested in continuous culture. The 2-λm derived replicative system could not be maintained in cells over high generation numbers. In continuous culture, the instability was more pronounced at high dilution rates (D) and high histidine concentration, for which the yeast is auxotrophic. The nature of the instability was probably due to a gene conversion event between the plasmid and the yeast chromosome. In contrast, the integrative expression system was stably maintained in cells over prolonged cultivation times. Since this work focused on the production of large quantities of P450 by heterologous expression in yeast over prolonged time periods, the integrant was used to optimize P450alk expression by varying continuous culture parameters. The P450alk expression was shown to be dependent on the D applied to the culture. The highest P450alk expression levels were obtained at high D, when cell metabolism was shifted to partial glucose oxidation, yielding ethanol as a major metabolite in the culture supernatant. In contrast, when glucose was completely oxidized at low D, the ADHI-dependent P450alk expression was reduced and followed by a corresponding decrease in heterologous protein.
Similar content being viewed by others
References
Ammerer G (1983) Expression of genes in yeast using the ADCI promoter. Methods Enzymol 101:192–202
Baldari C, Cesareni G (1985) Plasmids pEMBLY: new single-stranded shuttle vectors for the recovery and analysis of yeast DNA sequences. Gene 35:27–32
Bentley WE, Kompala DS (1989) A novel structured kinetic modeling approach for the analysis of plasmid instability in recombinant bacterial cultures. Biotechnol Bioeng 33:49–61
Bugeja VC, Kleinmann MJ, Stanbury PF, Gingold EB (1989) The segregation of the 2 λm-based yeast plasmid pJDB248 breaks down under conditions of slow, glucose-limited growth. J Gen Microbiol 135:2891–2897
Bühler M, Schindler M (1984) Aliphatic hydrocarbons. In: Kiesling K (ed) Biotechnology, vol 6a. Verlag Chemie, Basle, pp 329–385
Caulcott CA, Dunn A, Robertson HA, Cooper NS, Brown ME, Rhodes PM (1987) Investigation of the effect of growth environment on the stability of low-copy-number plasmids in Escherichia coli. J Gen Microbiol 133:1881–1889
Ciriacy M (1975) Genetics of alcohol dehydrogenase in Saccharomyces cerevisiae. Mutat Res 29:315–326
Clewell DB, Helinski DR (1969) Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an open circular DNA form. Proc Natl Acad Sci USA 62:1159–1166
Cohen SN, Chang ACY, Hsu L (1972) Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci USA 69:2110–2114
Cohen SN, Chang ACY (1973) Recircularization and autonomous replication of a sheared R-factor DNA segment in Escherichia coli transformants. Proc Natl Acad Sci USA 70:1293–1297
Denis CL, Ferguson J, Young ET (1983) mRNA levels for the fermentative alcohol dehydrogenase of Saccharomyces cerevisiae decrease upon growth on a nonfermentable carbon source. J Biol Chem 258:1165–1171
Dobson MJ, Kingsman SM, Kingsman AJ (1981) Sequence variation in the LEU2 region of the Saccharomyces cerevisiae genome. Gene 16:133–139
Futcher AB, Cox BS (1984) Copy number and the stability of 2-λm circle-based artificial plasmids of Saccharomyces cerevisiae. J Bacteriol 157:283–290
Hinnen A, Meyhack B (1982) Vectors for cloning in yeast. Curr Topics Microbiol Immunol 96:101–107
Hinnen A, Hicks HB, Fink GR (1978) Transformation of yeast. Proc Natl Acad Sci USA 75:1929–1933
Holm C, Meeks-Wagner DW, Fangman WL, Botstein D (1986) A rapid, efficient method for isolating DNA from yeast. Gene 42:169–173
Hug H, Blanch HW, Fiechter A (1974) The functional role of lipids in hydrocarbon assimilation. Biotechnol Bioeng 16:965–985
Impoolsup A, Caunt P, Greenfield PF (1989) Effect of growth rate on stability of a recombinant plasmid during continuous culture of Saccharomyces cerevisiae in non-selective medium. J Biotechnol 10:171–180
Kingsman AJ, Gimlich RL, Clarke L, Chinault AC, Carbon J (1981) Sequence variation in dispersed repetitive sequences in Saccharomyces cerevisiae. J Mol Biol 145:619–632
Kleinman MJ, Gingold EB, Stanbury PF (1986) The stability of the yeast plasmid pJDB248 depends on growth rate of the culture. Biotechnol Lett 8:225–230
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Lee F-JS, Hassan HM (1987) Effect of oxygen tension on stability and expression of a killer toxin chimeric plasmid in a chemostat culture of Saccharomyces cerevisiae. Appl Microbiol Biotechnol 27:72–74
Lee GM, Song KB, Rhee SK, Han MH (1986) Plasmid maintenance and growth of recombinant Saccharomyces cerevisiae producing hepatitis B virus surface antigen. Biotechnol Lett 8:385–390
Maniatis T, Fritsch EF, Sambrook J (1982) Molecular cloning. Cold Spring Harbor Laboratory, Cold Spring Harbor, N. Y.
Marquet M, Alouani S, Hass ML, Loison G, Brown SW (1987) Double mutants of Saccharomyces cerevisiae harbour stable plasmids: stable expression of a eukaryotic gene and the influence of host physiology during continuous culture. J Biotechnol 6:135–145
Mead DJ, Gardner DCJ, Oliver SG (1986) Enhanced stability of a 2 λm-based recombinant plasmid in diploid yeast. Biotechnol Lett 8:391–396
Omura T, Sato R (1964) The carbon monooxide-binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J Biol Chem 239:2370–2378
Parker C, DiBiasio D (1987) Effect of growth rate and expression level on plasmid stability in Saccharomyces cerevisiae. Biotechnol Bioeng 29:215–221
Petrick M, Käppeli O, Fiechter A (1983) An expanded concept for the glucose effect in the yeast Saccharomyces uvarum: involvement of short- and long-term regulation. J Gen Microbiol 129:43–49
Rothstein R (1985) Cloning in yeast. In: Glover DM (ed) DNA cloning, vol 2. IRL Press, Oxford, p 45
Sakaki T, Oeda K, Yabusaki Y, Ohkawa H (1986) Monooxygenase activity of Saccharomyces cerevisiae cells transformed with expression plasmids carrying rat cytochrome P-450MC cDNA. J Biochem 99:741–749
Sanglard D, Loper JC (1989) Characterization of the alkane-inducible cytochrome P450 (P450alk) gene from the yeast Candida tropicalis: identification of a new P450 gene family. Gene 76:121–136
Sanglard D, Beretta I, Wagner M, Käppeli O, Fiechter A (1990) Functional expression of the alkane-inducible monooxygenase system of the yeast Candida tropicalis in Saccharomyces cerevisiae. Biocatalysis 4:19–28
Schaffner W, Weissmann C (1973) A rapid, sensitive, and specific method for the determination of protein in dilute solution. Anal Biochem 56:502–514
Srienc F, Campbell JL, Bailey JE (1986) Analysis of unstable recombinant Saccharomyces cerevisiae population growth in selective medium. Biotechnol Bioeng 18:996–1006
Tornow J, Santangelo GM (1990) Efficient expression of the Saccharomyces cerevisiae glycolytic gene ADH1 is dependent upon a cis-acting regulatory element (UAS rpg) found initially in genes encoding ribosomal proteins. Gene 90:79–85
Weber JM, Reiser J, Käppeli O (1990) Lanosterol 14α-demethylase-encoding gene: systematic analysis of homologous over-expression in Saccharomyces cerevisiae using strong yeast promoters. Gene 87:167–175
Wiseman A, King DJ (1982) Microbial Oxygenases- and their potential applications. Top Enzyme Ferment Biotechnol 6:151–206
Young V, Williamson A, Taguchi A, Smith M, Sledziewski A, Russell D, Osterman J, Denis C, Cox D, Beier D (1982) The alcohol dehydrogenase genes of the yeast Saccharomyces cerevisiae: isolation, structure and regulation. In: Hollaender A (ed), Genetic engineering of microorganisms for chemicals. Plenum Press, New York, p 335
Zurbriggen B, Kühne AB, Kallio P, Käppeli O, Fiechter A (1989) Controlled expression of heterologous cytochrome P450e cDNA in Saccharomyces cerevisiae. J Biotechnol 9:273–286
Author information
Authors and Affiliations
Additional information
Offsprint requests to: D. Sanglard
Rights and permissions
About this article
Cite this article
Beretta, I., Sanglard, D., Käppeli, O. et al. Optimization of Candida tropicalis cytochrome P450alk gene expression in Saccharomyces cerevisiae with continuous cultures. Appl Microbiol Biotechnol 36, 48–60 (1991). https://doi.org/10.1007/BF00164698
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00164698