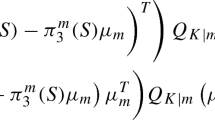Abstract
Many real problems in supervised classification involve high-dimensional feature data measured for individuals of known origin from two or more classes. When the dimension of the feature vector is very large relative to the number of individuals, it presents formidable challenges to construct a discriminant rule (classifier) for assigning an unclassified individual to one of the known classes. One way to handle this high-dimensional problem is to identify highly relevant differential features for constructing a classifier. Here a new approach is considered, where a mixture model with random effects is used firstly to partition the features into clusters and then the relevance of each feature variable for differentiating the classes is formally tested and ranked using cluster-specific contrasts of mixed effects. Finally, a non-parametric clustering approach is adopted to identify networks of differential features that are highly correlated. The method is illustrated using a publicly available data set in cancer research for the discovery of correlated biomarkers relevant to the cancer diagnosis and prognosis.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Benjamini, Y., Hochberg, Y.: Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 259–300 (1995)
Bickel, P.J., Levina, E.: Some theory for Fisher’s linear discriminant function, ‘naive Bayes’, and some alternatives when there are many more variables than observations. Bernoulli 10, 989–1010 (2004)
Borgatti, S.P., Everett, M.G., Freeman, L.C.: Ucinet for Windows: Software for Social Network Analysis. Analytic Technologies, Harvard, MA (2002). Available via http://www.analytictech.com/. Accessed 8 Dec 2015
Cai, T., Liu, W.: A direct estimation approach to sparse linear discriminant analysis. J. Am. Stat. Assoc. 106, 1566–1577 (2011)
Collado, M., Garcia, V., Garcia, J.M., Alonso, I., Lombardia, L., et al.: Genomic profiling of circulating plasma RNA for the analysis of cancer. Clin. Chem. 53, 1860–1863 (2007)
Dahl, D.B., Newton, M.A.: Multiple hypothesis testing by clustering treatment effects. J. Am. Stat. Assoc. 102, 517–526 (2007)
Donoho, D., Jin, J.: Higher criticism for large-scale inference, especially for rare and weak effects. Stat. Sci. 30, 1–25 (2015)
Fan, J., Lv, J.: A selective review of variable selection in high dimensional feature space. Stat. Sin. 20, 101–148 (2010)
Fan, J., Feng, Y., Tong, X.: A road to classification in high dimensional space: the regularized optimal affine discriminant. J. R. Stat. Soc. B 74, 745–771 (2012)
Hall, P., Pittelkow, Y., Ghosh, M.: Theoretic measures of relative performance of classifiers for high-dimensional data with small sample sizes. J. R. Stat. Soc. B 70, 158–173 (2008)
Hall, P., Jin, J., Miller, H.: Feature selection when there are many influential features. Bernoulli 20, 1647–1671 (2014)
He, Y., Pan, W., Lin, J.: Cluster analysis using multivariate normal mixture models to detect differential gene expression with microarray data. Comput. Stat. Data Anal. 51, 641–658 (2006)
Kersten, J.: Simultaneous feature selection and Gaussian mixture model estimation for supervised classification problems. Pattern Recogn. 47, 2582–2595 (2014)
Matsui, S., Noma, H.: Estimating effect sizes of differentially expressed genes for power and sample-size assessments in microarray experiments. Biometrics 67, 1225–1235 (2011)
McLachlan, G.J.: Discriminant analysis. WIREs Comput. Stat. 4, 421–431 (2012)
McLachlan, G.J., Peel, D.: Finite Mixture Models. Wiley, New York (2000)
McLachlan, G.J., Do, K.A., Ambroise, C.: Analyzing Microarray Gene Expression Data. Wiley, New York (2004)
Ng, S.K.: A two-way clustering framework to identify disparities in multimorbidity patterns of mental and physical health conditions among Australians. Stat. Med. 34, 3444–3460 (2015)
Ng, S.K., McLachlan, G.J.: Mixture models for clustering multilevel growth trajectories. Comput. Stat. Data Anal. 71, 43–51 (2014)
Ng, S.K., McLachlan, G.J., Wang, K., Ben-Tovim, L., Ng, S.-W.: A mixture model with random-effects components for clustering correlated gene-expression profiles. Bioinformatics 22, 1745–1752 (2006)
Ng, S.K., Holden, L., Sun, J.: Identifying comorbidity patterns of health conditions via cluster analysis of pairwise concordance statistics. Stat. Med. 31, 3393–3405 (2012)
Ng, S.K., McLachlan, G.J., Wang, K., Nagymanyoki, Z., Liu, S., Ng, S.-W.: Inference on differences between classes using cluster-specific contrasts of mixed effects. Biostatistics 16, 98–112 (2015)
Pan, W., Lin, J., Le, C.T.: Model-based cluster analysis of microarray gene-expression data. Genome Biol. 3, 0009.1–0009.8 (2002)
Pyne, S., Lee, S.X., Wang, K., Irish, J., Tamayo, P., et al.: Joint modeling and registration of cell populations in cohorts of high-dimensional flow cytometric data. PLoS One 9, e100334 (2014)
Qi, Y., Sun, H., Sun, Q., Pan, L.: Ranking analysis for identifying differentially expressed genes. Genomics 97, 326–329 (2011)
Qiu, W., He, W., Wang, X., Lazarus, R.: A marginal mixture model for selecting differentially expressed genes across two types of tissue samples. Int. J. Biostat. 4, Article 20 (2008)
Smyth, G.: Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, Article 3 (2004)
Storey, J.D.: The optimal discovery procedure: a new approach to simultaneous significance testing. J. R. Stat. Soc. B 69, 347–368 (2007)
Zhao, Y.: Posterior probability of discovery and expected rate of discovery for multiple hypothesis testing and high throughput assays. J. Am. Stat. Assoc. 106, 984–996 (2011)
Acknowledgements
Part of this work has been presented in the Conference of the International Federation of Classification Societies, Bologna, July 2015. This work was supported by a grant from the Australian Research Council.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Ng, S.K., McLachlan, G.J. (2017). On the Identification of Correlated Differential Features for Supervised Classification of High-Dimensional Data. In: Palumbo, F., Montanari, A., Vichi, M. (eds) Data Science . Studies in Classification, Data Analysis, and Knowledge Organization. Springer, Cham. https://doi.org/10.1007/978-3-319-55723-6_4
Download citation
DOI: https://doi.org/10.1007/978-3-319-55723-6_4
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-55722-9
Online ISBN: 978-3-319-55723-6
eBook Packages: Mathematics and StatisticsMathematics and Statistics (R0)