Abstract
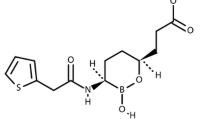
The structurally unrelated antimicrobials, macrolides, lincosamides, and streptogramins are grouped into a single family, called MLS family. This classification is based on a similar, although not identical, mechanism of action. Macrolides are composed of a minimum of two amino and/or neutral sugars attached to a lactone ring of variable size [1] (Fig. 18.1). Erythromycin, produced by a strain of the actinomycete Saccharopolyspora erythraea (formerly Streptomyces erythraeus), is the first macrolide discovered in 1952. It actually corresponds to a mixture of antibiotics that includes erythromycin A, which is the active compound and has a 14-membered lactone ring with two sugars, cladinose and an amino sugar (e.g., desosamine). Other commercially available macrolides derived from erythromycin A include clarithromycin, dirithromycin, roxithromycin, as well as azithromycin that has an enlarged 15-membered ring resulting from a nitrogen insertion. Structural modifications of erythromycin A resulted in improved pharmacokinetic profiles and better tolerance, but cross-resistance between members of this class of antimicrobials was still observed. Some 16-membered ring macrolides are also available in a few countries (spiramycin, josamycin, midecamycin, and miocamycin) or for veterinary use (tylosin). The most recent class of ketolides comprises telithromycin and cethromycin (ABT-773), which are derived from clarithromycin and have two major modifications, replacement of cladinose by a keto-function and an 11-12-carbamate extension with an alkyl-aryl modification in telithromycin. The first fluoroketolide solithromycin (CEM-101), exhibiting a different side chain and a fluorine atom linked to C-2 of the lactone, shows higher in vitro activity and enhanced accumulation in macrophages as compared to telithromycin [2].
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Takashima H. Structural consideration of macrolide antibiotics in relation to the ribosomal interaction and drug design. Curr Top Med Chem. 2003;3:991–9.
Llano-Sotelo B, Dunkle J, Klepacki D, Zhang W, Fernandes P, Cate JH, Mankin AS. Binding and action of CEM-101, a new fluoroketolide antibiotic that inhibits protein synthesis. Antimicrob Agents Chemother. 2010;54:4961–70.
Spizek J, Novotna J, Rezanka T. Lincosamides: chemical structure, biosynthesis, mechanism of action, resistance, and applications. Adv Appl Microbiol. 2004;56:121–54.
Mukhtar TA, Wright GD. Streptogramins, oxazolidinones, and other inhibitors of bacterial protein synthesis. Chem Rev. 2005;105:529–42.
Canu A, Leclercq R. Overcoming bacterial resistance by dual target inhibition: the case of streptogramins. Curr Drug Targets Infect Disord. 2001;1:215–25.
Barriere JC, Berthaud N, Beyer D, Dutka-Malen S, Paris JM, Desnottes JF. Recent developments in streptogramin research. Curr Pharm Des. 1998;4:155–80.
Johnston NJ, Mukhtar TA, Wright GD. Streptogramin antibiotics: mode of action and resistance. Curr Drug Targets. 2002;3:335–44.
Politano AD, Sawyer RG. NXL-103, a combination of flopristin and linopristin, for the potential treatment of bacterial infections including community-acquired pneumonia and MRSA. Curr Opin Investig Drugs. 2010;11:225–36.
Verdier L, Bertho G, Gharbi-Benarous J, Girault JP. Lincomycin and clindamycin conformations. A fragment shared by macrolides, ketolides and lincosamides determined from TRNOE ribosome-bound conformations. Bioorg Med Chem. 2000;8:1225–43.
Agmon I, Amit M, Auerbach T, Bashan A, Baram D, Bartels H, Berisio R, Greenberg I, Harms J, Hansen HA, Kessler M, Pyetan E, Schluenzen F, Sittner A, Yonath A, Zarivach R. Ribosomal crystallography: a flexible nucleotide anchoring tRNA translocation, facilitates peptide-bond formation, chirality discrimination and antibiotics synergism. FEBS Lett. 2004;567:20–6.
Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–20.
Yonath A. Ribosomal crystallography: peptide bond formation, chaperone assistance and antibiotics activity. Mol Cells. 2005;20:1–16.
Harms J, Schluenzen F, Zarivach R, Bashan A, Gat S, Agmon I, Bartels H, Franceschi F, Yonath A. High resolution structure of the large ribosomal subunit from a mesophilic Eubacterium. Cell. 2001;107:679–88.
Tu D, Blaha G, Moore PB, Steitz TA. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell. 2005;121:257–70.
Wilson DN, Harms JM, Nierhaus KH, Schlünzen F, Fucini P. Species-specific antibiotic-ribosome interactions: implications for drug development. Biol Chem. 2005;386:1239–52.
Schlünzen F, Zarivach R, Harms J, Bashan A, Tocilj A, Albrecht R, Yonath A, Franceschi F. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature. 2001;413:814–21.
Polacek N, Mankin AS. The ribosomal peptidyl transferase center: structure, function, evolution, inhibition. Crit Rev Biochem Mol Biol. 2005;40:285–311.
Poehlsgaard J, Douthwaite S. The bacterial ribosome as a target for antibiotics. Nat Rev Microbiol. 2005;13:870–81.
Gaynor M, Mankin AS. Macrolide antibiotics: binding site, mechanism of action, resistance. Curr Top Med Chem. 2003;3:949–60.
Yonath A, Bashan A. Ribosomal crystallography: initiation, peptide bond formation, and amino acid polymerization are hampered by antibiotics. Annu Rev Microbiol. 2004;58:233–51.
Cocito C, Di Giambattista M, Nyssen E, Vannuffel P. Inhibition of protein synthesis by streptogramins and related antibiotics. J Antimicrob Chemother. 1997;39(Suppl A):7–13.
Weisblum B. Erythromycin resistance by ribosome modification. Antimicrob Agents Chemother. 1995;39:577–85.
Liu M, Douthwaite S. Activity of the ketolide telithromycin is refractory to Erm monomethylation of bacterial rRNA. Antimicrob Agents Chemother. 2002;46:1629–33.
Douthwaite S, Jalava J, Jakobsen L. Ketolide resistance in Streptococcus pyogenes correlates with the degree of rRNA dimethylation by Erm. Mol Microbiol. 2005;58:613–22.
Leclercq R, Courvalin P. Resistance to macrolides and related antibiotics in Streptococcus pneumoniae. Antimicrob Agents Chemother. 2002;46:2727–34.
Weisblum B. Insights into erythromycin action from studies of its activity as inducer of resistance. Antimicrob Agents Chemother. 1995;39:797–805.
Tanaka T, Weisblum B. Mutant of Staphylococcus aureus with lincomycin- and carbomycin-inducible resistance to erythromycin. Antimicrob Agents Chemother. 1974;5:538–40.
Clarebout G, Nativelle E, Leclercq R. Unusual inducible cross resistance to macrolides, lincosamides, and streptogramins B by methylase production in clinical isolates of Staphylococcus aureus. Microb Drug Resist. 2001;7:317–22.
Rosato A, Vicarini H, Bonnefoy A, Chantot JF, Leclercq R. A new ketolide, HMR 3004, active against streptococci inducibly resistant to erythromycin. Antimicrob Agents Chemother. 1998;42:1392–6.
Lewis II JS, Jorgensen JH. Inducible clindamycin resistance in staphylococci: should clinicians and microbiologists be concerned? Clin Infect Dis. 2005;40:280–5.
Fines M, Gueudin M, Ramon A, Leclercq R. In vitro selection of resistance to clindamycin related to alterations in the attenuator of the erm(TR) gene of Streptococcus pyogenes UCN1 inducibly resistant to erythromycin. J Antimicrob Chemother. 2001;48:411–6.
Rao GG. Should clindamycin be used in treatment of patients with infections caused by erythromycin-resistant staphylococci? J Antimicrob Chemother. 2000;45:715–6.
McGehee RF, Barrett FF, Finland F. Resistance of Staphylococcus aureus to lincomycin, clindamycin, and erythromycin. Antimicrob Agents Chemother. 1968;13:392–7.
Drinkovic D, Fuller ER, Shore KP, Holland DJ, Ellis-Pegler R. Clindamycin treatment of Staphylococcus aureus expressing inducible clindamycin resistance. J Antimicrob Chemother. 2001;48:315–6.
Frank AI, Marcinak JF, Mangat PD, Tjhio JT, Kelkar S, Schreckenberger PC, Quinn JP. Clindamycin treatment of methicillin-resistant Staphylococcus aureus infections in children. Pediatr Infect Dis J. 2002;21:530–4.
Siberry GK, Tekle T, Carroll K, Dick J. Failure of clindamycin treatment of methicillin-resistant Staphylococcus aureus expressing inducible clindamycin resistance in vitro. Clin Infect Dis. 2003;37:1257–60.
Levin TP, Suh B, Axelrod P, Truant AL, Fekete T. Potential clindamycin resistance in clindamycin-susceptible, erythromycin-resistant Staphylococcus aureus: report of a clinical failure. Antimicrob Agents Chemother. 2005;49:1222–4.
Daurel C, Huet C, Dhalluin A, Bes M, Etienne J, Leclercq R. Differences in potential for selection of clindamycin-resistant mutants between inducible erm(A) and erm(C) Staphylococcus aureus genes. J Clin Microbiol. 2008;46:546–50.
Clarebout G, Nativelle E, Bozdogan B, Villers C, Leclercq R. Bactericidal activity of quinupristin-dalfopristin against strains of Staphylococcus aureus with the MLS(B) phenotype of resistance according to the erm gene type. Int J Antimicrob Agents. 2004;24:444–9.
Schwarz S, Werckenthin C, Kehrenberg C. Identification of a plasmid-borne chloramphenicol-florfenicol resistance gene in Staphylococcus sciuri. Antimicrob Agents Chemother. 2000;44:2530–3.
Toh SM, Xiong L, Arias CA, Villegas MV, Lolans K, Quinn J, Mankin AS. Acquisition of a natural resistance gene renders a clinical strain of methicillin-resistant Staphylococcus aureus resistant to the synthetic antibiotic linezolid. Mol Microbiol. 2007;64:1506–14.
Witte W, Cuny C. Emergence and spread of cfr-mediated multiresistance in staphylococci: an interdisciplinary challenge. Future Microbiol. 2011;6:925–31.
Diaz L, Kiratisin P, Mendes RE, Panesso D, Singh KV, Arias CA. Transferable plasmid-mediated resistance to linezolid due to cfr in a human clinical isolate of Enterococcus faecalis. Antimicrob Agents Chemother. 2012;56:3917–22.
Kehrenberg C, Schwarz S, Jacobsen L, Hansen LH, Vester B. A new mechanism for chloramphenicol, florfenicol and clindamycin resistance: methylation of 23S ribosomal RNA at A2503. Mol Microbiol. 2005;57:1064–73.
Long KS, Poehlsgaard J, Kehrenberg C, Schwarz S, Vester B. The Cfr rRNA methyltransferase confers resistance to Phenicols, Lincosamides, Oxazolidinones, Pleuromutilins, and Streptogramin A antibiotics. Antimicrob Agents Chemother. 2006;50:2500–5.
Tait-Kamradt A, Davies T, Cronan M, Jacobs MR, Appelbaum PC, Sutcliffe J. Mutations in 23S rRNA and ribosomal protein L4 account for resistance in pneumococcal strains selected in vitro by macrolide passage. Antimicrob Agents Chemother. 2000;44:2118–25.
Roberts MC. Update on macrolide-lincosamide-streptogramin, ketolide, and oxazolidinone resistance genes. FEMS Microbiol Lett. 2008;282:147–59.
Arthur M, Andremont A, Courvalin P. Distribution of erythromycin esterase and rRNA methylase genes in members of the family Enterobacteriaceae highly resistant to erythromycin. Antimicrob Agents Chemother. 1987;3:404–9.
Barthélémy P, Autissier D, Gerbaud G, Courvalin P. Enzymic hydrolysis of erythromycin by a strain of Escherichia coli. A new mechanism of resistance. J Antibiot. 1984;37:1692–6.
Noguchi N, Emura A, Matsuyama H, O’Hara K, Sasatsu M, Kono M. Nucleotide sequence and characterization of erythromycin resistance determinant that encodes macrolide 2′-phosphotransferase I in Escherichia coli. Antimicrob Agents Chemother. 1995;39:2359–63.
Noguchi N, Katayama J, O’Hara K. Cloning and nucleotide sequence of the mphB gene for macrolide 2′-phosphotransferase II in Escherichia coli. FEMS Microbiol Lett. 1996;144:197–202.
Schmitz FJ, Sadurski R, Kray A, Boos M, Geisel R, Kohrer K, Verhoef J, Fluit AC. Prevalence of macrolide-resistance genes in Staphylococcus aureus and Enterococcus faecium isolates from 24 European university hospitals. J Antimicrob Chemother. 2000;45:891–4.
Chesneau O, Tsvetkova K, Courvalin P. Resistance phenotypes conferred by macrolide phosphotransferases. FEMS Microbiol Lett. 2007;269:317–22.
Boumghar-Bourtchai L, Mariani-Kurkdjian P, Bingen E, Filliol I, Dhalluin A, Ifrane SA, Weill FX, Leclercq R. Macrolide-resistant Shigella sonnei. Emerg Infect Dis. 2008;14:1297–9.
Phuc Nguyen MC, Woerther PL, Bouvet M, Andremont A, Leclercq R, Canu A. Escherichia coli as reservoir for macrolide resistance genes. Emerg Infect Dis. 2009;15:1648–50.
Matsuoka M, Endou K, Kobayashi H, Inoue M, Nakajima Y. A plasmid that encodes three genes for resistance to macrolide antibiotics in Staphylococcus aureus. FEMS Microbiol Lett. 1998;167:221–7.
Leclercq R, Brisson-Noel A, Duval J, Courvalin P. Phenotypic expression and genetic heterogeneity of lincosamides inactivation in Staphylococcus spp. Antimicrob Agents Chemother. 1987;31:1887–91.
Bozdogan B, Berrezouga L, Kuo M, Yurek D, Farley K, Stockman B, Leclercq R. A new resistance gene, linB, conferring resistance to lincosamides by nucleotidylation in Enterococcus faecium HM1025. Antimicrob Agents Chemother. 1999;43:925–9.
Heir E, Lindstedt BA, Leegaard TM, Gjernes E, Kapperud G. Prevalence and characterisation of integrons in blood culture Enterobacteriaceae and gastrointestinal Escherichia coli in Norway and reporting of a novel class I integron-located lincosamide resistance gene. Ann Clin Microbiol Antimicrob. 2004;3:12.
Wang J, Shoemaker N, Wang GR, Salyers A. Characterization of a Bacteroides mobilizable transposon of a functional lincomycin resistance gene. J Bacteriol. 2000;182:3559–71.
Achard A, Villers C, Pichereau V, Leclercq R. New lnu(C) gene conferring resistance to lincomycin by nucleotidylation in Streptococcus agalactiae UCN36. Antimicrob Agents Chemother. 2005;49:2716–9.
Petinaki E, Guérin-Faublée V, Pichereau V, Villers C, Achard A, Malbruny B, Leclercq R. Lincomycin resistance gene lnu(D) in Streptococcus uberis. Antimicrob Agents Chemother. 2008;52:626–30.
Achard A, Leclercq R. Characterization of a small mobilizable transposon, MTnSag1, in Streptococcus agalactiae. J Bacteriol. 2007;189:4328–31.
Gravey F, Galopin S, Grall N, Auzou M, Andremont A, Leclercq R, Cattoir V. Lincosamide resistance mediated by lnu(C) (L phenotype) in a Streptococcus anginosus clinical isolate. J Antimicrob Chemother. 2013;68(11):2464–7.
Haenni M, Saras E, Bertin S, Leblond P, Madec JY, Payot S. Diversity and mobility of integrative and conjugative elements in bovine isolates of Streptococcus agalactiae, S. dysgalactiae subsp. dysgalactiae, and S. uberis. Appl Environ Microbiol. 2010;76:7957–65.
Haenni M, Saras E, Chaussière S, Treilles M, Madec JY. ermB-mediated erythromycin resistance in Streptococcus uberis from bovine mastitis. Vet J. 2011;189:356–8.
Hershberger E, Donabedian S, Konstantinou K, Zervos MJ. Quinupristin-dalfopristin resistance in gram-positive bacteria: mechanism of resistance and epidemiology. Clin Infect Dis. 2004;38:92–8.
Li XZ, Nikaido H. Efflux-mediated drug resistance in bacteria. Drugs. 2004;64:159–204.
Singh KV, Weinstock GM, Murray BE. An Enterococcus faecalis ABC homologue (Lsa) is required for the resistance of this species to clindamycin and quinupristin-dalfopristin. Antimicrob Agents Chemother. 2002;46:1845–50.
Kehrenberg C, Ojo KK, Schwarz S. Nucleotide sequence and organization of the multiresistance plasmid pSCFS1 from Staphylococcus sciuri. J Antimicrob Chemother. 2004;54:936–9.
Malbruny B, Werno AM, Anderson TP, Murdoch DR, Leclercq R. A new phenotype of resistance to lincosamides and streptogramin A-type antibiotics in Streptococcus agalactiae in New Zealand. J Antimicrob Chemother. 2004;54:1040–4.
Malbruny B, Werno AM, Murdoch DR, Leclercq R, Cattoir V. Cross-resistance to lincosamides, streptogramins A, and pleuromutilins due to the lsa(C) gene in Streptococcus agalactiae UCN70. Antimicrob Agents Chemother. 2011;55:1470–4.
Li B, Wendlandt S, Yao J, Liu Y, Zhang Q, Shi Z, Wei J, Shao D, Schwarz S, Wang S, Ma Z. Detection and new genetic environment of the pleuromutilin-lincosamide-streptogramin A resistance gene lsa(E) in methicillin-resistant Staphylococcus aureus of swine origin. J Antimicrob Chemother. 2013;68:1251–5.
Dowzicky M, Talbot GH, Feger C, Prokocimer P, Etienne J, Leclercq R. Characterization of isolates associated with emerging resistance to quinupristin/dalfopristin (Synercid) during a worldwide clinical program. Diagn Microbiol Infect Dis. 2000;37:57–62.
Isnard C, Malbruny B, Leclercq R, Cattoir V. Genetic basis for in vitro and in vivo resistance to lincosamides, streptogramins A and pleuromutilins (LSAP phenotype) in Enterococcus faecium. Antimicrob Agents Chemother. 2013;57(9):4463–9.
Haroche J, Morvan A, Davi M, Allignet J, Bimet F, El Solh N. Clonal diversity among streptogramin A-resistant Staphylococcus aureus isolates collected in French hospitals. J Clin Microbiol. 2003;41:586–91.
Novotna G, Janata J. A new evolutionary variant of the streptogramin A resistance protein, Vga(A)LC, from Staphylococcus haemolyticus with shifted substrate specificity towards lincosamides. Antimicrob Agents Chemother. 2006;50:4070–6.
Tessé S, Trueba F, Berthet N, Hot C, Chesneau O. Resistance genes underlying the LSA phenotype of French staphylococcal isolates. Antimicrob Agents Chemother. 2013;57(9):4543–6.
Ambrose KD, Nisbet R, Stephens DS. Macrolide efflux in Streptococcus pneumoniae is mediated by a dual efflux pump (mel and mef) and is erythromycin inducible. Antimicrob Agents Chemother. 2005;48:4203–9.
Leclercq R. Mechanisms of resistance to macrolides and lincosamides: nature of the resistance elements and their clinical implications. Clin Infect Dis. 2002;34:482–92.
Santagati M, Iannelli F, Oggioni MR, Stefani S, Pozzi G. Characterization of a genetic element carrying the macrolide efflux gene mef(A) in Streptococcus pneumoniae. Antimicrob Agents Chemother. 2000;44:2585–7.
Gay K, Stephens DS. Structure and dissemination of a chromosomal insertion element encoding macrolide efflux in Streptococcus pneumoniae. J Infect Dis. 2001;184:56–65.
Ojo KK, Striplin MJ, Ulep CC, Close NS, Zittle J, Luis H, Bernardo M, Leitao J, Roberts MC. Staphylococcus efflux msr(A) gene characterized in Streptococcus, Enterococcus, Corynebacterium, and Pseudomonas isolates. Antimicrob Agents Chemother. 2006;50:1089–91.
Davidson AL, Dassa E, Orelle C, Chen J. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol Mol Biol Rev. 2008;72:317–64.
Ross JI, Eady EA, Cove JH, Cunliffe WJ, Baumberg S, Wootton JC. Inducible erythromycin resistance in staphylococci is encoded by a member of the ATP-binding transport super-gene family. Mol Microbiol. 1990;4:1207–14.
Reynolds E, Ross JI, Cove JH. Msr(A) and related macrolide/streptogramin resistance determinants: incomplete transporters? Int J Antimicrob Agents. 2003;22:228–36.
Fiebelkorn KR, Crawford SA, McElmeel ML, Jorgensen JH. Practical disk diffusion method for detection of inducible clindamycin resistance in Staphylococcus aureus and coagulase-negative staphylococci. J Clin Microbiol. 2003;41:4740–4.
Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing; 21th informational supplement. M100-S11. Wayne, PA: CLSI; 2011.
Jorgensen JH, Crawford SA, McElmeel ML, Fiebelkorn KR. Detection of inducible clindamycin resistance of staphylococci in conjunction with performance of automated broth susceptibility testing. J Clin Microbiol. 2004;42:1800–2.
Zelazny AM, Ferraro MJ, Glennen A, Hindler JF, Mann LM, Munro S, Murray PR, Reller LB, Tenover FC, Jorgensen JH. Selection of strains for quality assessment of the disk induction method for detection of inducible clindamycin resistance in staphylococci: a CLSI collaborative study. J Clin Microbiol. 2005;43:2613–5.
Burucoa C, Garnier M, Silvain C, Fauchère JL. Quadruplex real-time PCR assay using allele-specific scorpion primers for detection of mutations conferring clarithromycin resistance to Helicobacter pylori. J Clin Microbiol. 2008;46:2320–6.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this chapter
Cite this chapter
Cattoir, V., Leclercq, R. (2017). Resistance to Macrolides, Lincosamides, and Streptogramins. In: Mayers, D., Sobel, J., Ouellette, M., Kaye, K., Marchaim, D. (eds) Antimicrobial Drug Resistance. Springer, Cham. https://doi.org/10.1007/978-3-319-46718-4_18
Download citation
DOI: https://doi.org/10.1007/978-3-319-46718-4_18
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-46716-0
Online ISBN: 978-3-319-46718-4
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)