Abstract
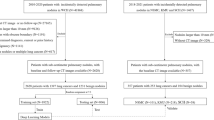
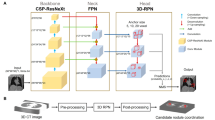
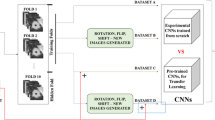
Diagnosis of pulmonary lesions from computed tomography (CT) is important but challenging for clinical decision making in lung cancer related diseases. Deep learning has achieved great success in computer aided diagnosis (CADx) area for lung cancer, whereas it suffers from label ambiguity due to the difficulty in the radiological diagnosis. Considering that invasive pathological analysis serves as the clinical golden standard of lung cancer diagnosis, in this study, we solve the label ambiguity issue via a large-scale radio-pathomics dataset containing 5,134 radiological CT images with pathologically confirmed labels, including cancers (e.g., invasive/non-invasive adenocarcinoma, squamous carcinoma) and non-cancer diseases (e.g., tuberculosis, hamartoma). This retrospective dataset, named Pulmonary-RadPath, enables development and validation of accurate deep learning systems to predict invasive pathological labels with a non-invasive procedure, i.e., radiological CT scans. A three-level hierarchical classification system for pulmonary lesions is developed, which covers most diseases in cancer-related diagnosis. We explore several techniques for hierarchical classification on this dataset, and propose a Leaky Dense Hierarchy approach with proven effectiveness in experiments. Our study significantly outperforms prior arts in terms of data scales (\(6\times \) larger), disease comprehensiveness and hierarchies. The promising results suggest the potentials to facilitate precision medicine.
J. Yang and M. Gao—These authors have contributed equally.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
Notes
- 1.
Note that the AUC metrics could not directly compare with each other since the datasets, inclusion criteria, and experiment settings are different.
References
Armato, S.G., et al.: The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI): a completed reference database of lung nodules on CT scans. Med. Phys. 38(2), 915–931 (2011)
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R.L., Torre, L.A., Jemal, A.: Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68(6), 394–424 (2018)
Cui, Y., Jia, M., Lin, T.Y., Song, Y., Belongie, S.J.: Class-balanced loss based on effective number of samples. In: CVPR, pp. 9260–9269 (2019)
Dou, Q., Chen, H., Jin, Y., Lin, H., Qin, J., Heng, P.-A.: Automated pulmonary nodule detection via 3D ConvNets with online sample filtering and hybrid-loss residual learning. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D.L., Duchesne, S. (eds.) MICCAI 2017. LNCS, vol. 10435, pp. 630–638. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-66179-7_72
Esteva, A., et al.: Dermatologist-level classification of skin cancer with deep neural networks. Nature 542, 115–118 (2017)
Finn, C., Abbeel, P., Levine, S.: Model-agnostic meta-learning for fast adaptation of deep networks. In: ICML (2017)
Gneiting, T., Raftery, A.E.: Strictly proper scoring rules, prediction, and estimation. J. Am. Stat. Assoc. 102, 359–378 (2007)
Gong, J., et al.: A deep residual learning network for predicting lung adenocarcinoma manifesting as ground-glass nodule on CT images. Eur. Radiol. 30, 1847–1855 (2019). https://doi.org/10.1007/s00330-019-06533-w
Guan, M.Y., Gulshan, V., Dai, A.M., Hinton, G.E.: Who said what: modeling individual labelers improves classification. In: AAAI (2017)
Guo, C., Pleiss, G., Sun, Y., Weinberger, K.Q.: On calibration of modern neural networks. In: ICML (2017)
Huang, G., Liu, Z., Van Der Maaten, L., Weinberger, K.Q.: Densely connected convolutional networks. In: CVPR, vol. 1, p. 3 (2017)
Huang, X., Yang, J., et al.: Evaluating and boosting uncertainty quantification in classification. arXiv preprint arXiv:1909.06030 (2019)
Hussein, S., Cao, K., Song, Q., Bagci, U.: Risk stratification of lung nodules using 3D CNN-based multi-task learning. In: Niethammer, M., et al. (eds.) IPMI 2017. LNCS, vol. 10265, pp. 249–260. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-59050-9_20
Ioffe, S., Szegedy, C.: Batch normalization: accelerating deep network training by reducing internal covariate shift. In: ICML (2015)
Kim, H., et al.: CT-based deep learning model to differentiate invasive pulmonary adenocarcinomas appearing as subsolid nodules among surgical candidates: comparison of the diagnostic performance with a size-based logistic model and radiologists. Eur. Radiol. 30(6), 3295–3305 (2020). https://doi.org/10.1007/s00330-019-06628-4
Kingma, D.P., Ba, J.: Adam: a method for stochastic optimization. arXiv preprint arXiv:1412.6980 (2014)
Lee, K., Lee, K., Min, K., Zhang, Y., Shin, J., Lee, H.: Hierarchical novelty detection for visual object recognition. In: CVPR, pp. 1034–1042 (2018)
Paszke, A., et al.: Automatic differentiation in PyTorch (2017)
Tang, H., Zhang, C., Xie, X.: NoduleNet: decoupled false positive reduction for pulmonary nodule detection and segmentation. In: Shen, D., et al. (eds.) MICCAI 2019. LNCS, vol. 11769, pp. 266–274. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-32226-7_30
Travis, W.D., et al.: The 2015 world health organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J. Thorac. Oncol. 10(9), 1243–1260 (2015)
Wang, S., et al.: Predicting EGFR mutation status in lung adenocarcinoma on computed tomography image using deep learning. Eur. Respir. J. 53, 1–11 (2019)
Xie, Y., Xia, Y., Zhang, J., Feng, D.D., Fulham, M., Cai, W.: Transferable multi-model ensemble for benign-malignant lung nodule classification on chest CT. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D.L., Duchesne, S. (eds.) MICCAI 2017. LNCS, vol. 10435, pp. 656–664. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-66179-7_75
Yang, J., Deng, H., Huang, X., Ni, B., Xu, Y.: Relational learning between multiple pulmonary nodules via deep set attention transformers. In: ISBI (2020)
Yang, J., Fang, R., Ni, B., Li, Y., Xu, Y., Li, L.: Probabilistic radiomics: ambiguous diagnosis with controllable shape analysis. In: Shen, D., et al. (eds.) MICCAI 2019. LNCS, vol. 11769, pp. 658–666. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-32226-7_73
Yang, J., Huang, X., Ni, B., Xu, J., Yang, C., Xu, G.: Reinventing 2D convolutions for 3D medical images. arXiv preprint arXiv:1911.10477 (2019)
Zhang, H., et al.: Context encoding for semantic segmentation. In: CVPR, June 2018
Zhao, W., et al.: Toward automatic prediction of EGFR mutation status in pulmonary adenocarcinoma with 3D deep learning. Cancer Med. 8, 3532–3543 (2019)
Zhao, W., et al.: 3D deep learning from CT scans predicts tumor invasiveness of subcentimeter pulmonary adenocarcinomas. Cancer Res. 78(24), 6881–6889 (2018)
Zhao, W., et al.: Convolution kernel and iterative reconstruction affect the diagnostic performance of radiomics and deep learning in lung adenocarcinoma pathological subtypes. Thorac. Cancer 10(10), 1893–1903 (2019)
Acknowledgment
This work was supported by National Science Foundation of China (61976137, U1611461). This work was also supported by Shanghai Municipal Health Commission (2018ZHYL0102, 2019SY072, 201940018). Authors appreciate the Student Innovation Center of SJTU for providing GPUs.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this paper
Cite this paper
Yang, J. et al. (2020). Hierarchical Classification of Pulmonary Lesions: A Large-Scale Radio-Pathomics Study. In: Martel, A.L., et al. Medical Image Computing and Computer Assisted Intervention – MICCAI 2020. MICCAI 2020. Lecture Notes in Computer Science(), vol 12266. Springer, Cham. https://doi.org/10.1007/978-3-030-59725-2_48
Download citation
DOI: https://doi.org/10.1007/978-3-030-59725-2_48
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-59724-5
Online ISBN: 978-3-030-59725-2
eBook Packages: Computer ScienceComputer Science (R0)