Abstract
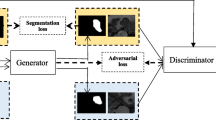
Semantic segmentation is an important task in medical image analysis. In general, training models with high performance needs a large amount of labeled data. However, collecting labeled data is typically difficult, especially for medical images. Several semi-supervised methods have been proposed to use unlabeled data to facilitate learning. Most of these methods use a self-training framework, in which the model cannot be well trained if the pseudo masks predicted by the model itself are of low quality. Co-training is another widely used semi-supervised method in medical image segmentation. It uses two models and makes them learn from each other. All these methods are not end-to-end. In this paper, we propose a novel end-to-end approach, called difference minimization network (DMNet), for semi-supervised semantic segmentation. To use unlabeled data, DMNet adopts two decoder branches and minimizes the difference between soft masks generated by the two decoders. In this manner, each decoder can learn under the supervision of the other decoder, thus they can be improved at the same time. Also, to make the model generalize better, we force the model to generate low-entropy masks on unlabeled data so the decision boundary of model lies in low-density regions. Meanwhile, adversarial training strategy is adopted to learn a discriminator which can encourage the model to generate more accurate masks. Experiments on a kidney tumor dataset and a brain tumor dataset show that our method can outperform the baselines, including both supervised and semi-supervised ones, to achieve the best performance.
This work is supported by the NSFC-NRF Joint Research Project (No. 61861146001).
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Badrinarayanan, V., Kendall, A., Cipolla, R.: SegNet: a deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 39(12), 2481–2495 (2017)
Bai, W., et al.: Semi-supervised learning for network-based cardiac MR image segmentation. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D.L., Duchesne, S. (eds.) MICCAI 2017. LNCS, vol. 10434, pp. 253–260. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-66185-8_29
Berthelot, D., Carlini, N., Goodfellow, I.J., Papernot, N., Oliver, A., Raffel, C.: MixMatch: a holistic approach to semi-supervised learning. CoRR (2019)
Blum, A., Mitchell, T.M.: Combining labeled and unlabeled data with co-training. In: Proceedings of Annual Conference on Computational Learning Theory (COLT) (1998)
Chartsias, A., et al.: Factorised spatial representation learning: application in semi-supervised myocardial segmentation. In: Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds.) MICCAI 2018. LNCS, vol. 11071, pp. 490–498. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00934-2_55
Chen, L., Papandreou, G., Kokkinos, I., Murphy, K., Yuille, A.L.: Semantic image segmentation with deep convolutional nets and fully connected CRFs. In: Proceedings of International Conference on Learning Representations (ICLR) (2015)
Chen, L., Zhu, Y., Papandreou, G., Schroff, F., Adam, H.: Encoder-decoder with atrous separable convolution for semantic image segmentation. In: Proceedings of European Conference on Computer Vision (ECCV) (2018)
Goodfellow, I.J., et al.: Generative adversarial nets. In: Proceedings of Neural Information Processing Systems (NIPS) (2014)
Han, B., Yao, Q., Yu, X., Niu, G., Xu, M., Hu, W., Tsang, I.W., Sugiyama, M.: Co-teaching: robust training of deep neural networks with extremely noisy labels. In: Proceedings of Neural Information Processing Systems (NIPS) (2018)
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: Proceedings of Computer Vision and Pattern Recognition (CVPR) (2016)
Jaccard, P.: Étude comparative de la distribution florale dans une portion des alpes et des jura. Bull. Soc. Vaudoise Sci. Nat. 37, 547–579 (1901)
Long, J., Shelhamer, E., Darrell, T.: Fully convolutional networks for semantic segmentation. In: Proceeding of Computer Vision and Pattern Recognition (CVPR) (2015)
Milletari, F., Navab, N., Ahmadi, S.: V-net: fully convolutional neural networks for volumetric medical image segmentation. In: Proceeding of 3D Vision (3DV) (2016)
Nie, D., Gao, Y., Wang, L., Shen, D.: ASDNet: attention based semi-supervised deep networks for medical image segmentation. In: Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds.) MICCAI 2018. LNCS, vol. 11073, pp. 370–378. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00937-3_43
Qiao, S., Shen, W., Zhang, Z., Wang, B., Yuille, A.L.: Deep co-training for semi-supervised image recognition. In: Proceedings of European Conference on Computer Vision (ECCV) (2018)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Souly, N., Spampinato, C., Shah, M.: Semi and weakly supervised semantic segmentation using generative adversarial network. CoRR (2017)
Zhou, Y., et al.: Collaborative learning of semi-supervised segmentation and classification for medical images. In: Proceeding of Computer Vision and Pattern Recognition (CVPR) (2019)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this paper
Cite this paper
Fang, K., Li, WJ. (2020). DMNet: Difference Minimization Network for Semi-supervised Segmentation in Medical Images. In: Martel, A.L., et al. Medical Image Computing and Computer Assisted Intervention – MICCAI 2020. MICCAI 2020. Lecture Notes in Computer Science(), vol 12261. Springer, Cham. https://doi.org/10.1007/978-3-030-59710-8_52
Download citation
DOI: https://doi.org/10.1007/978-3-030-59710-8_52
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-59709-2
Online ISBN: 978-3-030-59710-8
eBook Packages: Computer ScienceComputer Science (R0)