Abstract
LifeWatch Italy, the Italian node of LifeWatch ERIC, has promoted and stimulated the debate on the use of semantics in biodiversity data management. Actually, biodiversity and ecosystems data are very heterogeneous and need to be better managed to improve the actual scientific knowledge extracted, as well as to address the urgent societal challenges concerning environmental issues. LifeWatch Italy has realized the Phytoplankton Virtual Research Environment (hereafter Phytoplankton VRE), a collaborative working environment supporting researchers to address basic and applied studies on phytoplankton ecology. The Phytoplankton VRE provides the IT infrastructure to enable researchers to obtain, share and analyse phytoplankton data at a level of resolution from individual cells to whole assemblages. A semantic approach has been used to address data harmonisation, integration and discovery: an interdisciplinary team has developed a thesaurus on phytoplankton functional traits and linked its concepts to other existing conceptual schemas related to the specific domain.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Phytoplankton plays an important role in aquatic ecosystems because it accounts for most global primary production and affects biogeochemical processes, trophic dynamics and biodiversity architecture. In order to understand ecosystem function and to improve predictions of aquatic ecosystem responses to environmental and climate change, it is strictly important that plankton physiologists and ecologists understand the phytoplankton structure.
In this chapter, we present the Phytoplankton Virtual Research Environment (Phytoplankton VRE), a collaborative working environment aimed at supporting researchers in addressing basic and applied studies on phytoplankton ecology. In particular, it allows researchers to analyse and share phytoplankton data at different resolutions: from individual cells to whole assemblages, data which are generally unharmonised and unavailable as online services. Moreover, it allows researchers to assess phytoplankton cell size, i.e. the biovolume, and other morphological traits.
The remainder of this chapter is organised as follows: Sect. 2 describes the architectural overview of the VRE, mainly based on a set of virtual machines that are accessible through a remote desktop connection. Section 3 is for the Phytoplankton case study and, in particular, it presents the important role played by Phytoplankton in aquatic environments, its main characteristics in terms of size, shape and other morphological traits, and the problem caused by the presence of several diverse methods to compute biovolume, surface area and other indices that do not allow researchers to compare data. The Phytoplankton VRE is then presented with all its tools and services: the atlas of taxonomy (Atlas of Phytoplankton), the atlas of morphological traits (Atlas of Shapes), the Phytoplankton Traits Thesaurus, and the Taverna workflow management system, that represents an orchestrator able to run all services composed in the workflow aimed at computing hidden dimension, biovolume, surface area, multi-metric indices of the ecological status, etc. The data lifecycle is also presented in order to illustrate all the stages involved in the management of data for their use and re-use (data acquisition, data curation, data access and data processing). Section 4 concludes the chapter.
2 The LifeWatch Italy Approach to VRE
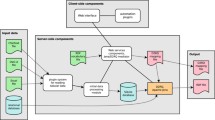
The Italian community that works on biodiversity and ecosystem research topics is composed for the most part by a multitude of little research groups. Investigators, usually, work with the limited resources of their laboratory: they can count on a laptop, equipped with computational and storage capabilities. In conceiving the architecture of the LifeWatch Italy VRE, we took into account that there was a resistance to changing the proper way investigators are expected to work. In order to reduce the resistance to change, we tried to maintain how investigators already work while supplying them more innovative services and unique storage and computational powers. The result is an architecture that supplies to researchers a set of Virtual Machines that are accessible through a Remote Desktop Connection very similar to the environment they normally use, but able to ensure very good performance in terms of computational capabilities and storage space, with no need to install additional tools on their workspace. Figure 1 shows an overview of the architecture. The requested Virtual Machine can be equipped in different ways, in line with the researchers’ goals. We give the possibility to set up the environment with an open source or Microsoft Operating System (i.e. Ubuntu or Windows Server), with all the tools needed for the data collection, curation and analysis. There is a Broker machine that is dedicated to managing all researchers’ connections and assigning dynamically computational and storage resources to users.
Each user has a dedicated account; a common Authentication and Authorization Infrastructure (AAI) based on Windows Active Directory is used to control the access and to assign different authorizations and rights on the machines. This allows users to have a personal desktop/area on each machine where they can organize their work in term of personal folders where they could store documents and files coming from the analysis done in the environment or shared folders if they need to share the work with other members in the LifeWatch network. The computational capacity of the environment is supplied by the distributed national data centres, but this is transparent to users that can just start their analysis without being worried about connection problems or being connected during analysis time. All analysis processes are run in the background and when they come back in the environment, they will find the results stored in the chosen folders.
An example of the Virtual Machine and User Desktop is introduced and described referring to the Study Case Study in the following sections.
3 The Phytoplankton Case Study
3.1 Overview
Phytoplankton is the primary autotrophic component in aquatic ecosystems, responsible for almost half of global net primary production [1]. On the one hand, it plays an important role in carbon sequestration and on the other hand, oxygen production. For this reason, this photosynthetic organism plays a key role in aquatic environments, forming the base of the food web and having a substantial function in nutrient dynamics and in the carbon biogeochemical cycle [2,3,4]. Therefore, considering the total phytoplankton structure, the community of phytoplankton has profound effects on higher trophic levels and key biogeochemical processes [5]. For these reasons, understanding both the role of phytoplankton features, traits and the abiotic and biotic drivers that determine phytoplankton distribution and its succession patterns is fundamental. The most important features of the phytoplankton community, organism size and elemental composition, will influence processes at the level of individuals, populations, communities and ecosystems [6, 7].
Size, shape, morphology and specific traits of these organisms provide relevant proxies for the ability to survive and coexist in response to abiotic and biotic drivers [8]. Regarding size, phytoplankton is an extremely diverse group of organisms that range over nine orders of magnitude in cell size volume and shapes [9, 10]. They show a huge scale of size from 1–2 µm in equivalent spherical diameter for the picoplankton, 2–20 µm for the nanoplankton, 20–200 µm for the microplankton, and up to 200–2000 µm for macroplankton [11, 12]. Every single phytoplankton organism is characterized also by a specific geometric shape. Currently, a well-defined number of shapes include simple and combined shapes (see Fig. 2) [13,14,15] that represent another morphological trait, very useful to describe and characterize phytoplankton community [16].
The Atlas of shapes. (The Atlas of shapes - Phytoplankton Bio-Imaging by the Ecology Unit of the University of Salento http://phytobioimaging.unisalento.it/Products/AtlasOfShapes.aspx?ID_Tipo=0)
Cell size is often referred to as a master trait because body size influences the physiology, ecology, and evolution of species [17]. Phytoplankton cell size varies over three orders of magnitude [10] and is mechanistically linked to all the physiological and ecological traits: maximum growth rate, nutrient acquisition, minimum and maximum cell quota, light absorption and susceptibility to high light stress, sinking rate and susceptibility to grazing and viral attack [10]. The size, structure and elemental composition of the phytoplankton community has a cascading influence on the proportion of organic material transferred to the microbial loop, higher trophic levels or exported into the deep sea. Phytoplankton size spectra and size classes have been shown to have high information content to detect environmental condition change in transitional and coastal waters [18,19,20].
It is therefore essential to investigate the development of phytoplankton populations in order to understand the biological functioning of aquatic systems and detect changes in them [21].
The physiological responses induced by different cell sizes and surface areas could provide valuable information about phytoplankton distributions as well as how distributions might be altered by environmental change. Cell size and shape may be the primary drivers of variations in physiological responses and therefore provide community assemblages with the flexibility to respond from macro spatial scale to local environmental conditions [8].
Regarding the other most important morphological trait, the shape, it provides important information about essential functional processes and ecological characteristic of phytoplankton. The geometric shape is traditionally used to calculate phytoplankton cell measurements (e.g. biovolume), but it can also play an important role in determining community distributions [22]. The geometric shape represents an interesting feature to be considered in the increasingly used trait-based approaches to the study and prediction of phytoplankton dynamics in aquatic ecosystems. The shape is easily observable and measurable and its application in the functional approach does not require taxonomic affiliation, although it provides important information about essential functional processes and ecological characteristics of phytoplankton organisms [22,23,24].
The high morphological variability of phytoplankton in terms of a geometric shape is not random. It is likely related to phytoplankton morphological adaptations to achieve the best fit with environmental conditions [22]. Based on shape, morphologically-based classifications for phytoplankton communities have been proposed by Stanca et al. in 2013 [22], an approach referred to as Phytoplankton Geometric Shapes (PGS). Phytoplankton species were allocated to the most similar geometric shape selected from those described by [9, 13] and [14]. At the same time, morphometric measurements (surface, volume and surface to volume ratios) were obtained from basic linear dimensions.
For plankton physiologists and ecologists, it is fundamental to understand phytoplankton structure in order to improve predictions of aquatic ecosystem responses to environmental and climate change. In addition, from a practical point of view and in the context of conservation, protection and management of aquatic resources, the assessment of phytoplankton community structure is essential to understand ecosystem function [25].
Due to their short life cycle, planktonic algae respond quickly to environmental changes. Therefore, phytoplankton is considered a useful biological quality element for water quality monitoring assessment, according to the European Water Framework Directive (WFD). Phytoplankton parameters to be used for this assessment are biomass, community composition and abundance, as well as frequency and intensity of blooms. This quality element responds mainly to pressures generated by nutrient and organic enrichment and alteration of the water body’s hydrological and morphological characteristics, to environmental forcing and to human-generated pressures [26].
Even though demographic traits (e.g. presence/absence, abundance and biomass) have been traditionally included in directives and monitoring programmes, morphological traits are attracting growing interest to be implemented as a descriptor of the ecological status of aquatic ecosystems [27,28,29]. Direct counts and measurements of algal size, in terms of biovolume, are potentially a more accurate measure of phytoplankton biomass and abundance [30]. Assessing phytoplankton cell size, i.e. the biovolume, has therefore been approached with different procedures and methodologies, each of which has aspects that need consideration and improvement.
The variety of applied methods, from sampling to counting, as well as the mode of calculation, unfortunately leads to general poor comparability of the data, which currently represents a huge problem. Indeed, to be shared and comparable, data have to answer to several data quality criteria. For this reason, they have to be sufficiently precise, accurate, representative and complete. Standardising protocols for validating and reporting data improves the comparability of data and the confidence with which one data set can be compared to another, either overtime or between research groups [31].
High-quality data is a key element for research and impacts the replicability of results. Quality checks should be performed during collection, data entry and analysis [32]. There have been many individual phytoplankton datasets collected across the world, but most of them are unavailable to the research community. The cornerstone action is to bring together data and information in a way that enables researchers to produce knowledge that yields novel insights or explanations, establishes correlations and identifies patterns [33]. Given the scale and urgency of the societal challenges related to the environment and given that data are being generated at an ever-increasing rate, better-coordinated efforts are required to enable structuring, aggregating, linking and processing of such data in a meaningful way [34]. Since quality assurance of data is an important component of the monitoring programme, the use of a standardised nomenclature list and a standardised computational model are decisive in improving the quality of the phytoplankton data and the comparability of results, at different spatial and temporal scales.
3.1.1 Comparability of Data: Taxonomy
New phytoplankton organisms are continuously being described, and changes in the naming and categorisation of organisms is common. Changes should be based on internationally accepted rules, which have been established in nomenclatural codes. It is essential to keep standardised lists, which are updated in a systematic way. Due to the inherent complexity of taxonomic, nomenclatural and systematic concepts, the quality and resolution of data are necessarily required [34]. For this reason and to optimize the management and integration of primary biodiversity data, it is necessary to develop a consistent vocabulary, semantic rules and ontologies; contribute to the harmonisation of terminology and practices; provide a synthetic guide for taxonomists and non-taxonomists involved in biomonitoring and biodiversity studies [34]. Moreover, when data from different sources, geographical areas and points in time are integrated into taxonomic inventories and databases or time series, they need a very careful critical revision, with the aim of internal consistency and quality evaluation [35] ensuring data interoperability and automated processing.
In order to do that, the “LifeWatch Taxonomic Backbone” service, a central part of the European LifeWatch Infrastructure set-up by the Flanders Marine Institute, can be exploited since it aims to (virtually) bring together different component databases and data systems, all of them related to taxonomy, biogeography, ecology, genetics and literature. By doing so, it standardises species data and integrates biodiversity data from different repositories.
3.1.2 Comparability of Data: Morphological Traits
Terminological ambiguity slows down scientific progress, leads to redundant research efforts, and ultimately impedes advances towards a unified foundation for ecological science [36, 37]. An important step of improvement of the phytoplankton analysis is the development of standard calculation procedures. There exists no unique procedure applied worldwide for all steps of phytoplankton morphological traits computation, no common set of protocols from linear dimensions measurement to biovolume calculation, which would allow inter-comparisons of data. Many countries or institutes have used their own methods for decades and may be reluctant to make changes [35]. Different measures and methodologies are in use to quantify cell size and they require unequivocal definition to ensure standardisation and comparability of measurements [37].
With the aim to solve the ambiguity issues of natural language by formalizing the construction of the terms themselves, their definitions and their inter-relationships, and in order to provide a standard set of structures that enable computers to more precisely assist data users in locating (data discovery) and processing the data of interest, we developed a specific thesaurus: the PhytoTraits thesaurus [37]. This thesaurus contains 120 terms hierarchically organized and focusing on morphofunctional traits, such as linear dimensions and shapes, which are univocally defined.
This controlled vocabulary provides a standard terminology for traits, that is essential for data integration and increasingly required in ecology. PhytoTraits is freely availableFootnote 1 and can be used for different purposes.
3.1.3 Comparability of Data: Computation Processes
Biovolume estimates and conversion factors required by indirect methods increase opportunities for error because the error associated with multiple independent factors can be propagated at each stage of calculation [38, 39].
The biovolume of phytoplankton must be assessed accurately in order to identify the ecological status of water bodies in line with the WFD requirements. There are several ways to calculate cell volumes. The ‘gold standard’ is to determine the geometrical shape that approximates the shape of the cell and then make measurements of the dimensions to enter into the formula for that particular geometrical shape [40,41,42,43]. Some of the challenges in this approach are that different investigators may choose a different geometric shape than the recommended shape [9, 14] for the same species, especially for cells with a complex shape. In addition, the ‘hidden dimension’ (i.e. the depth dimension) is difficult to measure since cells are viewed in two dimensions under the microscope [44].
Evaluating the most exact cell biovolume should help to avoid errors such as an overestimation or underestimation of phytoplankton biomass/biovolume. A properly estimated biovolume based on verified and agreed geometric shapes should lead to an accurate and comparable assessment of a phytoplankton-based ecological status [45].
To facilitate and accelerate the estimations of phytoplankton biovolume, which has become very important in the WFD-required ecological status assessment of water bodies, we revised and rearranged basic geometric shapes. Moreover, we verified and improved the precision as well as the accuracy of different formulas. Since only up to two dimensions can be visualised under the microscope, at least one dimension has to be derived from one of the others or a fixed value has to be determined from a number of specimens in a special effort. We calculated and provided conversion factors, hidden dimension factors, which are species-specific, in order to obtain dimensions that are difficult to measure, but needed for biovolume calculation. In this way, we provided a more accurate biovolume calculation, at a specific taxonomic level. We provided a set of 51 geometric models, including formulas for biovolume assessment and cell linear dimensions evaluation. There are two typology groups: Simple shape and Complex shape, with 23 and 28 shapes respectively. The models are provided in a specific Atlas, but also in a specific workflow developed for biovolume computation.
Having the opportunity to be more accurate and doing massive computation analysis allows for the reduction of mistakes and errors due to manual procedures and operator, permits the saving of time and should contribute to having fast answers in evaluating the ecological status of water bodies and providing more accurate results in line with the WFD requirements.
3.2 The Phytoplankton Virtual Research Environment
The e-Biodiversity Research Institute of LifeWatch Italy (hereafter LW ITA) has realized the Phytoplankton Virtual Research Environment (hereafter Phytoplankton VRE), a collaborative working environment supporting researchers to address basic and applied studies on phytoplankton ecology. The Phytoplankton VRE provides the IT infrastructure to enable researchers to obtain, share and analyse phytoplankton data at a level of resolution from individual cells to whole assemblages. The Phytoplankton VRE allows researchers to:
-
1.
Obtain and share harmonised data on taxonomy and morphological traits by using the Atlas of Phytoplankton, the Atlas of Shapes and the Phytoplankton Traits Thesaurus.
-
2.
Discover, access, integrate and export both own and other datasets (including additional metadata) held by LifeWatch Data Portal or distributed data centres.
-
3.
Share and create workflows by means of orchestrators like Taverna WorkbenchFootnote 2 by using algorithms and web services.
-
4.
Work together in a real-time environment that fosters the sharing of knowledge overcoming the limitations of traditional working practices e.g. the transfer of large datasets among users or the need for significant computational power for the analysis.
3.2.1 Harmonised Data on Taxonomy and Morphological Traits
The Phytoplankton VRE provides a number of features for harmonising phytoplankton taxonomic data and morphological trait data:
-
The Atlas of Phytoplankton: this provides a reference point for marine, transitional and freshwater scientists and students involved in phytoplankton identification and classification. It includes illustrative cards with information about i) taxonomy, with pictures and schematic drawings, information on similar species and/or synonyms, references; ii) ecological characteristics and geographical distribution of species; and iii) morphological features, such as shape association, linear dimensions association and formulae for cell volume and surface computation.
-
The Atlas of Shapes: this represents a reference point for marine, transitional and freshwater scientists and students involved in phytoplankton morphological traits association and measurement and provides a schematic protocol for calculating biovolume of phytoplankton species detectable with the Utermöhl method [46] in transitional ecosystems of the different world ecoregions. The Atlas includes the illustrative scheme of the shape classification subdivided in “Simple Shapes” and “Complex Shapes” (Fig. 2). Clicking on a specific shape, users are able to see: the biovolume (V) and surface area (A) computational models; and the shape views (e.g., lateral, frontal, etc.) with the corresponding linear dimensions (e.g. length indicated by alphabetical code “a”, “l”, etc.; width indicated by alphabetical code “b”, “d”, etc.). For each specific shape group there is the frontal view for the shape and the biovolume and area computational models. Clicking again on a specific shape, user is redirected to all taxonomic cards characterised by the selected shape that are on the Atlas of Phytoplankton. Both atlases are integrated and can be easily browsed, switching from taxonomic identification to morphological characterisation of phytoplankton.
-
The Phytoplankton Traits Thesaurus (PhytoTraits): this thesaurus reflects the agreement of a scientific expert community regarding the definition of semantic properties of approximately 120 traits [37]. Following Semantic Web standard technologies, the thesaurus has been implemented in Simple Knowledge Organisation System (SKOS), a common data model based on the Resource Description Framework (RDF). The PhytoTraits is freely available onlineFootnote 3, it can be queried through a SPARQL endpointFootnote 4 and is also accessible via APIFootnote 5 for integration with other systems. If adopted as a standard, and hence rigorously applied and enriched by the scientific community, PhytoTraits has the potential to significantly reduce the barriers to data discovery, integration, and exchange since it provides harmonised concepts with associated unique and resolvable URIs.
3.2.2 Data Access, Discovery, Integration and Download
A user who is registered at the LifeWatch Data Portal can access their own section titled “My Datasets” that lists all types of datasets in which he/she is involved (e.g. enabled, pending, refused, disabled or owned by users). For a specific dataset, the user can perform two main actions: download the dataset (RDF or CSV format) and/or visualize it in a separate window in JSON format.
Data Search Interface: this interface allows researchers to search species according to several dimensions of analysis. The searching criteria are: the geographic area (by drawing a polygon or a circle on the map); the biogeographic regions; the country; the ecosystem type; the habitat type; the organism group; and the scientific name. The resulting datasets are characterised by a title and an author. They are enriched with metadata and the user has to request authorization in order to access them. An advanced data search can be performed after that the administrator gives access to the dataset.
3.2.3 Sharing and Creating Workflows
The Phytoplankton VRE allows researchers to use Taverna, a workbench for the design and execution of scientific workflows. This tool enables the interoperation among databases and tools by providing a toolkit for composing, executing and managing workflow experiments. Taverna Workbench Biodiversity is an edition of Taverna that includes support for building and executing scientific workflows targeting biodiversity services. Taverna workflows show intermediate results of the execution, are easy to use for inexperienced users, and very flexible for the skilled ones. Taverna Workbench Biodiversity allows the use of a set of local and remote services to analyse and manage data, create nested workflows and use automatic iteration.
In order to facilitate the computation of phytoplankton traits and to investigate their distribution patterns, we developed a workflow, which allows automating a set of operations that were originally written in the R languageFootnote 6. Two R scripts have been developed and are incorporated in the PhytoTraitsComputationAndDistribution workflow:
-
the Phytoplankton Traits Computation, which computes morphological and demographic traits, such as hidden dimension, biovolume, surface area, surface-volume ratio, cell carbon content, density, carbon content and total biovolume;
-
the Phytoplankton Size Distributions, which performs Modality (Hartigans’ dip test), Normality or LogNormality (Anderson-Darling test, Cramer-von Mises) tests of phytoplankton biovolume (expressed as μm3) or cell carbon content (expressed as pgC*cell-1) distributions, at different levels of data aggregation (i.e. spatial, temporal, taxonomic).
The workflow is represented in Fig. 3. By default, “input ports” are shown on the top, and “output ports” are shown on the bottom. Boxes represent processing nodes, and the solid directed arrows between them are data connections. User has to specify for each input port:
-
1.
CompTraits: the input port for entering traits to be computed. Users shall select the “add value” button and enter one or more of these options in the box to compute: Biovolume; Surface Area (typing SA); Surface/Volume ratio (typing SV); Cells/Liter (typing CL); Biovolume/Liter (typing BVL); Carbon content (typing CC); Carbon content/Liter (typing CCL).
-
2.
PhytoRawData: the input port for entering raw data in CSV format. The workflow runs only with data resources structured according to the LifeWatch Italy Data Schema.
-
3.
CalcType: the input port for entering the computation type. Users can choose between two computation modalities:
-
a.
“Simplified” mode approximates the taxa specific-biovolume calculation/computation based on two linear dimensions only, length and width. The mandatory fields for this calculation/computation type are scientific name, measurement remarks (e.g. vision of the organism, dimension more or less than 20 μm), length and width;
-
b.
“Advanced” mode allows a more accurate estimate of taxon-specific biovolume, but it requires more information.
-
a.
For each shape, at least two measured basic linear dimensions need to be provided by the user. The mandatory fields for this calculation/computation type are scientific name, measurement remarks and linear dimensions. The latter must be measured according to the Phytobioimaging Atlas of Shapes.
-
4.
Cluster: the input port for entering the level of aggregation for size distributions. The aggregation could be done at spatial (e.g. eventid, paraeventid, locality, country and Eunis habitat type-name), and/or temporal level (e.g. day, month and year), and/or using taxonomic categories (e.g. Phylum, Order and scientific name). Size distributions will be aggregated according to a unique combination of the provided criteria (e.g. using three countries, four eventids for country and three dates for eventid as aggregation criteria, users will aggregate data in a single bin for each combination of country, eventid, and collection date, resulting in 3 × 4 × 3 = 36 bins).
-
5.
SizeUnit: the input port for entering the morphological trait that will be used to perform Modality, Normality or LogNormality tests of distributions.
-
6.
GraphicFileName: the input port for entering the name that will be used to create a PDF distribution file that will be visible on a web page once the workflow is completed.
Once the user has inserted the input values, the input dialogue window will close and users will be directed to the “results” screen, where it is possible to monitor the workflow execution progress in real-time. The first iteration of “PhytoTraitsComputation” will produce as output the dataset “TraitsOutput” in CSV format that contains all input data and computed traits, while the second iteration “PhytoSizeDistribution” will produce another CSV file “SizeDistributionOutput” reporting a summary of the distribution tests and “PhytoDistributionGraph”. At the end of the workflow process, users will automatically obtain also the distribution calculation graph.
3.3 Data Lifecycle
The data lifecycle illustrates the stages involved in the management of data for their use and re-use. There exists a wide range of data lifecycle models, each with a different focus or perspective. Starting from the DataONE Data Life Cycle framework [47] we customised the cycle according to our needs as represented in Fig. 4.
3.3.1 Data Acquisition
Data from the observations and experiments of individual investigators usually occur in heterogeneous formats and terminology (the same variable is reported with different terms or abbreviations) and are stored in flat files or spreadsheets with minimal formal structure and few or even without metadata information.
Data compilation is, therefore, an important and necessary step of the procedure for morphological and demographic traits calculation. We compiled a data template in which ancillary information (e.g. sampling site, longitude and latitude) and phytoplankton features (e.g. linear dimensions and surface area) are well-structured in a Microsoft Excel file. This procedure is important in order to import the file in the Phytoplankton Bio-Imaging System, to allow the traits calculation and to make the data interoperable.
The Excel file is structured in different fields related to a semantic model proposed by LW ITA (LifeWatch Data Management SystemFootnote 7), which clearly defines semantics that can be understood by researchers or interpreted by machines making it possible to determine appropriate uses of the data encoded therein.
According to this model, the ancillary data inherits concepts from the Darwin Core standardFootnote 8 and from the EnvThes vocabularyFootnote 9. The Darwin Core standard includes a glossary of terms, which aims to create a common language for sharing biodiversity primary data and related information, while EnvThes consists of lists of standardised terms for the description of data and information within geological, ecological and hydrological sciences. Data regarding phytoplankton traits are related to the Phytoplankton Traits Thesaurus (described in 3.1.2), which is a hierarchical controlled vocabulary designed to define a set of key terms and to capture how they are associated with each other in order to standardise scientific data on phytoplankton functional traits and to facilitate the access and exchange of information [37]. The LW ITA Semantic Model provides the relevant meta-information about the dataset fields (e.g., Name, Description, Data Type, Unit of Measure, Standard etc.) by solving ambiguities associated with data markup and also enabling records to be interpreted by computers.
The data acquisition step represents the data entry stage. Researchers can upload their own files to be shared, that can be in three main forms: a Comma-Separated values (CSV) file, a Darwin Core (DwC) file, an Access to Biological Collections Data (ABCD) document.
3.3.2 Data Curation
A culture of data curation and sharing is only recently establishing itself in ecology and new tools are needed to collect, harmonise, store, share and analyse ecological data. In this context, the use of computer automation to control the quality and consistency of data is of great help in identifying numerical or lexical inconsistencies within data strings coming from assembling different datasets. Computer automation may be also applied to check for the inevitable human mistakes that an operator, who has to insert hundreds or thousands of individual records, can commit. An example in this case study is a short code used to check for disproportionate numerical values that may potentially arise from typos or inconsistency of units of measurements.
The collection, correction and harmonisation of data is only a first step towards understanding the investigated ecosystem or biological processes investigated. The data curation stage is to guarantee accuracy and to assess the quality and includes all steps required to clean and validate data uploaded by researchers that have to comply with FAIR (Findability, Accessibility, Interoperability, and Reusability) principle [48] for sharing purposes. This stage includes 4 sub-stages:
-
Data assurance: the quality of data is assured by means of checks, inspections and validation procedures that allow to detect format errors, nomenclatural errors, numeric warnings, taxonomic warnings, and semantic warnings. Data assurance is performed by means of different automatic or semi-automatic tools available on the LifeWatch Data Portal.
-
Data description: data are accurately described by using an appropriate metadata standard to ensure understanding and long-term control. The LW Data Model gives the relevant meta information about the dataset fields (e.g., Name, Description, Data Type, Unit of measure, etc.).
-
Data deposit: data are published and hence made available to other researchers by providing adequate provenance (allowing to achieve authoring).
-
Data preservation: data are stored and preserved to be available in real-time for usage. This step allows also to manage and administer all curation lifecycle actions.
3.3.3 Data Access
Access control rules and authentication procedures are applied to ensure that only allowed users can access and use data. The technology used is Microsoft Active Directory.
-
Authentication: to access LW IT VRE, users need to have a LW VRE account. This account also gives access to the service catalogue and data resources.
-
Authorisation: first access to any Virtual Lab needs to be authorised by the LW administrator. Users will be “pending” until the administrator authorises their access. After that, users’ access will be enabled.
3.3.4 Data Processing
Ecologists collectively produce (and have historically produced) large volumes of data through diverse individual projects. Furthermore, the recent developments in Information and Communication Technologies give to ecologists the possibility to access two new types of data: new information created from new technology applications (e.g. remote sensing observations and advanced microscopy) and existing information that was previously unavailable (e.g. existent data that were not publicly available or simply not previously uploaded to an online source). It is difficult and often even impossible to characterise the functioning of a complex system by means of direct measurements. The size of the system and the complexity of the involved interactions often make necessary the use of descriptors able to summarise the collected information. In the case of large datasets, this need has to be extended also to the tools necessary to analyse data and produce summary indicators. Biomass, composition, abundance and size spectra of the phytoplankton community, as well as frequency and intensity of phytoplankton blooms, have been considered as fundamental summary descriptors to be included in the assessment of the aquatic ecological status. The data processing stage includes 3 sub-stages:
-
Data integration: data coming from heterogeneous data sources are combined in order to form homogeneous sets of data that can be easily and readily analysed.
-
Data discovery: data are provided to interested users for knowledge discovery purposes which are enriched with relevant and structured information (metadata).
-
Data analysis: data are explored and analysed by researchers according to the needs to create derived results useful for research, teaching and learning purposes.
Within the phytoplankton case study, we provided computational tools to calculate in a fast and automated way and at any chosen level of spatial and temporal aggregation:
-
The biovolume and biomass of any recorded individual cell starting from its linear dimensions measured at the microscope; considering the high variety of geometrical shapes that phytoplankton cells have, this tool is associated with a species-specific inventory of shapes and mathematical formulation needed for the calculation of the biovolume.
-
Different biodiversity indices, including taxonomic indices of richness, diversity, evenness and dominance and indices based on the size spectra of the phytoplanktonic community.
-
Inferences on the distribution of body mass across phytoplankton individuals (normality, log-normality, bimodality tests).
4 Conclusion
In this chapter we demonstrated how the LifeWatch Italy experience can be exploited by a group of researchers to address basic and applied studies on phytoplankton ecology. The Phyto VRE is able to reduce the chance of error and to optimise the whole process, the analysis and the processing and computational time. One of the challenges in computing the biovolume of a phytoplankton organism is represented by the fact that different investigators may choose a different geometric shape with respect to the recommended one for the same species, especially in the case of cells having a complex shape. The proposed Atlas of Shapes allows to have a reference point for marine, transitional and freshwater scientists and students interested in phytoplankton biodiversity and ecology. Moreover, the added value of the proposed approach is represented by the fact that it can be reproduced and exploited also for other studies (e.g. for alien species).
In conceiving the architecture of the proposed VRE, we considered the typical “resistance to change” of most researchers and we tried to maintain their way of working providing them innovative services and unique storage and computational powers. The result is an architecture that supplies to researchers a set of virtual machines that are accessible through a remote desktop connection that is very similar to the environment they normally use at work, but able to ensure very good performances in terms of computational capabilities and storage space. This is the main advantage of the proposed VRE with respect for convenience aspects, but it also represents the main limit of the proposed approach. As future work, we plan to provide web-based access to the VRE and hence to design and develop user-friendly interfaces able to answer to different users’ needs and expertise.
Notes
- 1.
PhytoTraits: http://thesauri.lifewatchitaly.eu/PhytoTraits/index.php.
- 2.
Taverna Workbench: www.taverna.org.uk.
- 3.
PythoTraits: http://thesauri.lifewatchitaly.eu/PhytoTraits/index.php.
- 4.
SPARQL endpoint: http://thesauri.lifewatchitaly.eu/PhytoTraits/sparql.php.
- 5.
PythoTraits API: http://thesauri.lifewatch.eu/PhytoTraits/services.php.
- 6.
R website: https://www.r-project.org.
- 7.
LifeWatch Data Management System: http://www.servicecentrelifewatch.eu/home.
- 8.
Darwin Core standard: http://rs.tdwg.org/dwc.
- 9.
EnvThes vocabulary: http://vocabs.ceh.ac.uk/evn/tbl/envthes.evn.
References
Field, C.B., Behrenfeld, M.J., Randerson, J.T., Falkowski, P.: Primary production of the biosphere: integrating terrestrial and oceanic components. Science 281(5374), 237–240 (1998)
Graham, L.E., Wilcox, L.W.: Algae. PrenticeHall, Upper Saddle River, New Jersey (2000)
Sarmiento, J.L., Gruber, N.: Ocean Biogeochemical Dynamics. Princeton University Press, Princeton (2006)
Almandoz, G.O., Hernando, M.P., Ferreyra, G.A., Schloss, I.R., Ferrario, M.E.: Seasonal phytoplankton dynamics in extreme southern South America (Beagle Channel, Argentina). J. Sea Res. 66, 47–57 (2011)
Litchman, E., Klausmeier, C.A., Schofield, O.M., Falkowski, P.G.: The role of functional traits and trade-offs in structuring phytoplankton communities: scaling from cellular to ecosystem level. Ecol. Lett. 10, 1170–1181 (2007)
Sterner, R.W., Elser, J.J.: Ecological Stoichiometry: The Biology of the Elements from Molecules to the Biosphere. Princeton University Press, Princeton (2002)
Hessen, D.O., Elser, J.J.: Elements of ecology and evolution. Oikos 109, 3–5 (2005)
Bestová, H., Munoz, F., Svoboda, P., Škaloud, P., Violle, C.: Ecological and biogeographical drivers of freshwater green algae biodiversity: from local communities to large - scale species pools of desmids. Oecologia 186, 1017–1030 (2018)
Hillebrand, H., Durselen, C.D., Kirschtel, D., Pollingher, U., Zohary, T.: Biovolume calculation for pelagic and benthic microalgae. J. Phycol. 35, 403–424 (1999)
Finkel, Z.V., Beardall, J., Flynn, K.J., Quigg, A., Rees, T.A.V., Raven, J.A.: Phytoplankton in a changing world: cell size and elemental stoichiometry. J. Plankton Res. 32, 119–137 (2010)
Sieburth, J.M., Smetacek, V., Lenz, J.: Pelagic ecosystem structure: heterotrophic compartments of the plankton and their relationship to plankton size fractions. Limnol. Oceanogr. 23, 1256–1263 (1978)
Beardall, J., et al.: Allometry and stoichiometry of unicellular, colonial and multicellular phytoplankton. New Phytol. 181(2), 295–309 (2009)
Vadrucci, M.R., Cabrini, M., Basset, A.: Biovolume determination of phytoplankton guilds in transitional water ecosystems of Mediterranean Ecoregion. Transit. Water. Bull. 2, 83–102 (2007)
Sun, J., Liu, D.Y.: Geometric models for calculating cell biovolume and surface area for phytoplankton. J. Plankton Res. 25, 1331–1346 (2003)
Atlas of Shapes - Phytoplankton Bio-Imaging by the Ecology Unit of the University of Salento. http://phytobioimaging.unisalento.it/en-us/products/AtlasOfShapes.aspx. Accessed 17 Dec 2019
Salmaso, N., Naselli-Flores, L., Padisak, J.: Functional classifications and their application in phytoplankton ecology. Freshw. Biol. 60, 603–619 (2015)
Finkel, Z.V., et al.: A universal driver of macroevolutionary change in the size of Marine phytoplankton over the Cenozoic. PNAS 104, 20416–20420 (2007)
Sabetta, L., Basset, A., Spezie, G.: Marine phytoplankton size–frequency distributions: spatial patterns and decoding mechanisms. Estuar. Coast. Shelf S. 80, 181–192 (2008)
Lugoli, F., et al.: Application of a new multi-metric phytoplankton index to the assessment of ecological status in marine and transitional waters. Ecol. Indic. 23, 338–355 (2012)
Vadrucci, M.R., et al.: Ability of phytoplankton trait sensitivity to highlight anthropogenic pressures in Mediterranean lagoons: a size spectra sensitivity index (ISS-phyto). Ecol. Indic. 34, 113–125 (2013)
Hötzel, G., Croome, R.: A phytoplankton methods manual for Australian Freshwaters. LWRRDC Occasional Paper 22/99. Land and Water Resources Research Development Corporation, Canberra, Australia (1999)
Stanca, E., Cellamare, M., Basset, A.: Geometric shape as a trait to study phytoplankton distributions in aquatic ecosystems. Hydrobiologia 701, 99–116 (2013)
Naselli-Flores, L., Padisák, J., Albay, M.: Shape and size in phytoplankton ecology: do they matter? Hydrobiologia 578, 157–161 (2007)
Salmaso, N., Padisák, J.: Morpho-functional groups and phytoplankton development in two deep lakes (Lake Garda, Italy and Lake Stechlin, Germany). Hydrobiologia 578, 97–112 (2007)
Choudhury, A.K., Bhadury, P.: Phytoplankton study from the Sundarbans ecoregion with an emphasis on cell biovolume estimates–a review. Indian J. Mar. Sci. 43(10), 1905–1913 (2014)
Varkitzi, I., et al.: Pelagic habitats in the Mediterranean Sea: a review of Good Environmental Status (GES) determination for plankton components and identification of gaps and priority needs to improve coherence for the MSFD implementation. Ecol. Indic. 95, 203–218 (2018)
Olenina, I., et al.: Biovolumes and size-classes of phytoplankton in the Baltic Sea HELCOM Balt. Sea Environ. Proc. n. 106 (2006)
OSPAR: OSPAR Integrated Report on the Eutrophication Status of the OSPAR Maritime Area Based Upon the First Application of the Comprehensive Procedure. Eutrophication Series. OSPAR Commission (2003)
HELCOM: Development of tools for assessment of eutrophication in the Baltic Sea. Baltic Sea Environmental Proceedings No. 104. Helsinki Commission (2006)
Carvalho, L., et al.: Strength and uncertainty of phytoplankton metrics for assessing eutrophication impacts in lakes. Hydrobiologia 704, 127–140 (2013)
King County: Marine Phytoplankton Monitoring Program Sampling and Analysis Plan. Prepared by A. Kolb, G. Hannach, L. Swanson, Water and Land Resources Division. Seattle, Washington (2016)
Sarmiento Soler, A., Ort, M., Steckel, J.: An Introduction to Data Management Reader_GFBio_BefMate_20160222, BEFmate, GFBio Project (2016)
Koureas, D., et al.: Unifying European biodiversity informatics. Res. Ideas Outcomes 2, e7787 (2016)
Sigovini, M., Keppel, E., Tagliapietra, D.: Open Nomenclature in the biodiversity era. Methods Ecol. Evol. 7, 1217–1225 (2016)
Zingone, A., et al.: Increasing the quality, comparability and accessibility of phytoplankton species composition time-series data. Estuar. CoastShelf S 162, 151–160 (2015)
Madin, J., Bowers, S., Schildhauer, M., Krivov, S., Pennington, D., Villa, F.: An ontology for describing and synthesizing ecological observation data. Ecol. Inform. 2, 279–296 (2007)
Rosati, I., et al.: A thesaurus for phytoplankton trait-based approaches: Development and applicability. Ecol. Inform. 42, 129–138 (2017)
Baguley, J.G., Hyde, L.J., Montagna, P.A.: A semi-automated digital microphotographic approach to measure meiofaunal biomass. Limnol. Oceanogr. Methods 2, 181–190 (2004)
Di Mauro, R., Cepeda, G., Capitanio, F., Viñas, M.D.: Using ZooImage automated system for the estimation of biovolume of copepods from the Northern Argentine Sea. J.- Sea Res. 66, 69–75 (2011)
Mullin, M.M., Sloan, P.R., Eppley, R.W.: Relationship between carbon content, cell volume, and area in phytoplankton. Limnol. Oceanogr. 11, 307–311 (1966)
Strathmann, R.R.: Estimating the organic carbon content of phytoplankton from cell volume or plasma volume. Limnol. Oceanogr. 12, 411–418 (1967)
Taguchi, S.: Relationships between photosynthesis and cell size of marine diatoms. J. Phycol. 12, 185–189 (1976)
Wheeler, P.A.: Cell geometry revisited: realistic shapes and accurate determination of cell volume and surface area from microscopic measurements. J. Phycol. 35, 209–210 (1999)
Harrison, P.J., et al.: Cell volumes of marine phytoplankton from globally distributed coastal data sets. Estuar. Coast. Shelf S. 162, 130–142 (2015)
Napiorkowska-Krzebietke, A., Kobos, J.: Assessment of the cell biovolume of phytoplankton widespread in coastal and inland water bodies. Water Res. 104, 532–546 (2016)
Edler, L., Elbrächter, M.: The Utermöhl method for quantitative phytoplankton analysis. In: Karlson, B., et al. (eds.) Microscopic and Molecular Methods for Quantitative Phytoplankton Analysis. Intergovernmental Oceanographic Commission Manuals and Guides 55, pp. 13–20. UNESCO, Paris (2010)
Michener, W.K., et al.: Participatory design of DataONE - Enabling cyberinfrastructure for the biological and environmental sciences. Ecol. Inform. 11, 5–15 (2012)
Wilkinson, M.D., Dumontier, M., Aalbersberg, I.J., et al.: The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 3, 160018 (2016). https://doi.org/10.1038/sdata.2016.18
Acknowledgements
This work was supported by the European Union’s Horizon 2020 research and innovation programme via the ENVRIplus project under grant agreement No 654182. The work was also funded by LifeWatch Italy, the Italian node of the escience European infrastructure for biodiversity and ecosystem research and also supported by “POR PUGLIA Progetto Strategico 2009–2012” (grant agreement CIP.PS-126).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2020 The Author(s)
About this chapter
Cite this chapter
Stanca, E., Fiore, N., Rosati, I., Vaira, L., Cozzoli, F., Basset, A. (2020). Case Study: LifeWatch Italy Phytoplankton VRE. In: Zhao, Z., Hellström, M. (eds) Towards Interoperable Research Infrastructures for Environmental and Earth Sciences. Lecture Notes in Computer Science(), vol 12003. Springer, Cham. https://doi.org/10.1007/978-3-030-52829-4_18
Download citation
DOI: https://doi.org/10.1007/978-3-030-52829-4_18
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-52828-7
Online ISBN: 978-3-030-52829-4
eBook Packages: Computer ScienceComputer Science (R0)