Abstract
Accurate registration of CT and CBCT images is key for adaptive radiotherapy. A particular challenge is the alignment of flexible organs, such as bladder or rectum, that often yield extreme deformations. In this work we analyze the impact of so-called structure guidance for learning based registration when additional segmentation information is provided to a neural network. We present a novel weakly supervised deep learning based method for multi-modal 3D deformable CT-CBCT registration with structure guidance constraints. Our method is not supervised by ground-truth deformations and we use the energy functional of a variational registration approach as loss for training. Incorporating structure guidance constraints in our learning based approach results in an average Dice score of \(0.91\pm 0.08\) compared to a score of \(0.76\pm 0.15\) for the same method without constraints. An iterative registration approach with structure guidance results in a comparable average Dice score of \(0.91\pm 0.09\). However, learning based registration requires only a single pass through the network, yielding computation of a deformation fields in less than 0.1 s which is more than 100 times faster than the runtime of iterative registration.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Deformable image registration (DIR) is an important tool in radiotherapy for cancer treatment. It is used for the alignment of a baseline CT and daily low-radiation cone beam CT (CBCT) images, allowing for motion correction, propagation of Hounsfield units and applied doses. Furthermore, organ segmentations, that are typically created by clinical experts during planning phase from the baseline CT, can be propagated to daily CBCT images. DIR has become a method of choice in image-guided radiotherapy and treatment planning over the last decades [2]. However, it is a demanding task that holds several challenges such as meaningful measurement of multi-modal similarity of CT and CBCT images, having low contrast and containing artifacts. Aside from that, flexible organs, such as bladder or rectum, can introduce extreme deformations, complicating an accurate registration. Conventional DIR algorithms such as [11] tend to underestimate large deformations, which is why extended DIR approaches were presented [9, 14]. These so-called structure guided approaches include information about corresponding anatomical delineations on the images in order to guide the registration. While the required delineations are usually available on the planning CT due to the workflow of radiotherapy, they need to be generated on CBCT scans before registration. As the advancements of machine learning algorithms proceed, fast and accurate generation of organ segmentations becomes easier, enabling structure guided DIR and making adaptive radiotherapy more feasible. However, DIR in radiotherapy remains a challenging task.
In the last few years, novel deep learning based registration methods have been proposed [12], showing potential of being superior to state-of-the-art iterative algorithms both in terms of accuracy and execution time. However, in the field of DIR in radiotherapy rather little work on deep learning based approaches has been done. In [3] for example a patch-based learning method for mono-modal CT-CT image registration has been proposed. Moreover, deep learning is used to overcome multi-modality by estimation of synthetic CT images from other modalities which then are used for registration [6]. As ground-truth deformations between images are hard to obtain, mostly unsupervised learning methods for DIR have been proposed in the past. Therefore a deep network is trained by minimization of a loss function inspired by the cost function of iterative registration methods [7, 15]. To include additional available information, such as organ delineations during training, so-called weakly supervised methods have been proposed and showed improved registration accuracy [1, 8]. Also in the context of radiotherapy these methods show promising results [10].
In this work we aim to combine the strengths of learning DIR with weak supervision and conventional registration using structure guidance. To this end we present a novel weakly supervised deep learning based method for multi-modal 3D deformable CT-CBCT registration with structure guidance constraints. Our method is not supervised by hard to obtain ground-truth deformation vector fields. The minimized loss is inspired by variational structure guided DIR algorithms, including an image similarity measure suitable for multi-modal CT-CBCT alignment and an additional term rating the alignment of given segmentation masks. Furthermore, we penalize deformation Jacobians to avoid local changes of topology and foldings. In contrast to existing learning based approaches, here we directly incorporate information on guidance structures as additional input to the networks. We evaluate our method on follow-up image pairs of the female pelvis and compare our results to conventional iterative registration algorithms.
Overview on our network training process. We train 3 different types of networks which all require the input of a reference and a template image. Additionally they can receive segmentations on the reference image or corresponding segmentations on both images as input (indicated by red dotted lines). The output is a deformation vector field that is applied to the template image and segmentations. The network parameters are updated using backpropagation based on the loss function presented in Sect. 2.2. (Color figure online)
2 Method
The goal of DIR is the generation of dense correspondences between a reference image \(\mathcal {R}\) and a template image \(\mathcal {T}\) with \(\mathcal {R},\mathcal {T}:\mathbb {R}^3 \rightarrow \mathbb {R}\). This is achieved by estimating a reasonable deformation vector field \(y:\varOmega \rightarrow \mathbb {R}^3\) on the field of view \(\varOmega \subseteq \mathbb {R}^3\) of \(\mathcal {R}\), such that the warped template image \(\mathcal {T}(y)\) and \(\mathcal {R}\) are similar. In a variational approach y is computed by minimizing a suitable cost function, usually consisting of an image similarity measure and a regularization term. In iterative registration this is typically done by a time-consuming gradient or Newton-type optimization scheme. However, in a deep learning based registration, the deformation is modeled by a convolutional neural network (CNN), that directly maps given input images to a vector field and that is parameterized with learnable parameters \(\theta \), i.e. \(y \equiv y_\theta (\mathcal {R},\mathcal {T})\). Due to the lack of ground-truth deformations, we adapt the variational approach and minimize the variational costs in average over all given training samples. In the context of learning, the cost function is the so-called loss function. An overview on the training process of our networks is given in Fig. 1.
2.1 Registration Types by Input
The networks require the input of a reference and a template image which need to be registered. Furthermore, we allow that available segmentations are provided as additional inputs. In this work, we distinguish between no additional input, a set \(\varSigma _\mathcal {R}=\{\varSigma _\mathcal {R}^\ell \subset \mathbb {R}^3, \ell =1,\ldots ,L\}\) of segmentations \(\varSigma _\mathcal {R}^\ell \) on the reference image, or two sets \(\varSigma _\mathcal {R}\), \(\varSigma _\mathcal {T}\) with corresponding segmentations \(\varSigma _\mathcal {R}^\ell \) and \(\varSigma _\mathcal {T}^\ell \) on reference and template image, respectively. On that account, we consider following three types of CNNs that predict a deformation field y depending on the given inputs:
-
Type I: \(y\equiv y_\theta (\mathcal {R},\mathcal {T})\) (images only)
-
Type II: \(y\equiv y_\theta (\mathcal {R},\mathcal {T},\varSigma _\mathcal {R})\) (images + reference segmentations)
-
Type III: \(y\equiv y_\theta (\mathcal {R},\mathcal {T},\varSigma _\mathcal {R},\varSigma _\mathcal {T})\) (images + corresponding segmentations)
Note that all three CNN registration types use information about anatomical structures during training for weak supervision. For inference, only the respective network inputs are required. Once the training process is finished, only a single pass though the network is needed for registration of unseen image pairs.
Above classification is clearly not limited to deep learning based registration as the registration types just describe the given inputs. In our experiments we will also refer to iterative registration of type I and III in analogues manner indicating the provided inputs per registration.
2.2 Loss Function
The loss function our networks minimize is similar to cost functions in iterative registration schemes [14]. It is composed of four parts, weighted by factors \(\alpha , \beta , \gamma > 0\):
with the edge-based normalized gradient fields (NGF) [5] distance measure
where \(\langle x,y\rangle _\varepsilon := x^\top y+\varepsilon \), \(\Vert x\Vert _\varepsilon := \sqrt{\langle x,x\rangle _{\varepsilon ^2}}\). Additionally, a \(L_2\)-penalty for weakly supervised structure guidance constraints is applied to segmentation masks hat are handled as multi-channel binary images \(\mathcal {M}_\mathcal {R},\mathcal {M}_\mathcal {T}:\mathbb {R}^3\rightarrow \{0,1\}^L\), such that \(\mathcal {M}_\mathcal {R}(x)_\ell =1\) iff \(x\in \varSigma _\mathcal {R}^\ell \) and \(\mathcal {M}_\mathcal {T}(x)_\ell =1\) iff \(x\in \varSigma _\mathcal {T}^\ell \). A spatial second order curvature regularization [4], where \(\varDelta y\equiv (\varDelta y_1, \varDelta y_2, \varDelta y_3)\) is the vector Laplacian, i.e. the Laplacian is applied component-wise, and a change of volume penalty with \(\psi (t):=(t-1)^2/t\) if \(t>0\) and \(\psi (t)=\infty \) otherwise are utilized to force physically reasonable deformations. The latter term penalizes Jacobians that indicate high volume growth \((\det \nabla y>1)\), shrinkage \((0<\det \nabla y<1)\) and especially unwanted grid foldings \((\det \nabla y\le 0)\).
2.3 Network Architecture
Our proposed CNN architecture is based on a U-Net [13] with four stages. Inputs are two 3D images \(\mathcal {R}\) and \(\mathcal {T}\) and, depending on the registration type (c.f. Sect. 2.1), additional reference segmentations \(\varSigma _\mathcal {R}\) or corresponding segmentations \(\varSigma _\mathcal {R}\) and \(\varSigma _\mathcal {T}\). Note that for each type a separate network has to be trained. First, individual convolution kernels are applied to each input. The results are combined by concatenation and afterwards convolution blocks and \(2\times 2\times 2\) max-pooling layers alternate. An convolution block consists of two convolutions with a kernel size of \(3\times 3\times 3\), each followed by a ReLU and a batch normalization layer. In each stage the number of feature channels gets doubled. In the decoder path, we alternate between transposed convolutions, convolution blocks and concatenating skip connections. Finally, we apply a \(1\times 1\times 1\) convolution, yielding the 3-channel deformation field with the same resolution as the inputs.
3 Experiments
We evaluate our proposed deep learning based method on image data of 31 female patients from multiple clinical sites. The dataset includes one planning CT and up to 26 follow-up CBCT scans of the pelvis for each patient, yielding 256 intrapatiental CT-CBCT image pairs in total. In order to focus on deformable parts of the registration, the images were affinely registered beforehand. Additionally the images were cropped to the same field of view and resampled to a size of \(160\times 160\times 80\) voxels, each with a size of approximately \(3\,\text {mm}\times 3\,\text {mm}\times 2\,\text {mm}\) in a preprocessing step. Available delineations of bladder, rectum and uterus were generated by clinical experts.
We evaluate the performance of three network types, differing in their number of required inputs and guidance through delineated structures. First, we only input two images that need to be registered. Second, we additionally include available segmentations on the reference CT image that are usually available after treatment planning phase. Third, we also include corresponding segmentations on the daily CBCT image for structure guidance. For comparison of our method with classical variational approaches we perform an iterative registration of all test image pairs, both with and without the guidance of given structures. We therefore minimize the same loss function without a volume change control term using an iterative L-BFGS optimizer. The weights in our loss and objective function (1), respectively, have been chosen manually as \(\alpha =30\), \(\beta =3\), \(\gamma =0.3\).
Each network type is evaluated performing a \(k-\)fold cross-validation with \(k=4\), splitting the dataset patient-wise into four subsets, training on three of them and testing on the left out subset. As evaluation measures we use the Dice similarity coefficient and the average surface distance (ASD) for estimation of segmentation overlap and registration accuracy. We check the plausibility of the deformation fields using their Jacobians as an indicator of volume changes and undesired grid foldings. The implementation of our deep learning framework is done using PyTorch and processed on a NVIDIA GeForce RTX 2070 with 8 GB memory and an Intel Core i7-9700K with 8 cores.
Comparison of Dice scores and average surface distances for all test images and annotated labels (bladder, rectum and uterus). For each label the distributions after the affine preregistration (
 ), a conventional iterative (
), a conventional iterative (
 ) and our proposed deep learning based registration (
) and our proposed deep learning based registration (
 ) are illustrated (c.f. Sect. 3). (Color figure online)
) are illustrated (c.f. Sect. 3). (Color figure online)
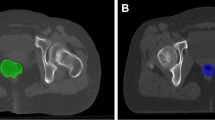
Qualitative comparison of registration results. (a) Reference and template with initial Dice score (average of the three scores for bladder, rectum, uterus). (b)-(f) Deformed template images \(\mathcal {T}(y)\) and deformations y for iterative (type I+III) and learning based DIR (type I-III). Additionally, we show segmentations of the bladder (
 ), rectum (
), rectum (
 ) and uterus (
) and uterus (
 ). (Color figure online)
). (Color figure online)
4 Results
The outcome of our experiments is summarized in Table 1. As expected, the registration quality improves with providing further input. We found that solely forwarding the reference and template image to our weakly supervised trained CNN for registration of type I yields an average Dice score of 0.76 (Dice after affine preregistration was 0.64). The result is superior to iterative registration of type I with an average Dice of 0.72. This is not surprising, as a particular advantage of learning based DIR algorithms is to build in anatomical knowledge and guidance, respectively, by weakly supervised training. Learning based registration of type II with additionally passing reference segmentations to the network slightly improves the registration accuracy (Dice 0.80), while providing corresponding segmentations on the CT and CBCT yields best results. In fact, structure guided iterative and learning based registration of type III both lead to an average Dice score of 0.91. Looking at the average surface distance shows a comparable tendency, where the iterative and learning based structure guided approaches both achieve values lower than the spatial resolution. The distributions of Dice scores and average surface distances are visualized in Fig. 3, showing a systematic improvement of registration results from type I to III.
A visual comparison of registration results of all types for one case is given in Fig. 4. Additionally, the results of structure guided iterative and learning based registrations of type III for three different patients are shown in Fig. 5. We observe that large deformations, especially of the bladder, are compensated due to the guidance of these structures. The plausibility of the underlying deformations can be checked with the help of the illustrated transformed grids. Furthermore, Fig. 2 displays the distributions of Jacobians for all approaches. As expected, Jacobians are centered around 1.0 with small standard deviations.
As specified in Table 1, the computational runtime of our deep learning based registration is over 100 times faster than the (CPU based) iterative approaches due to the fact that registration only needs a single pass through the CNN.
5 Conclusion
We presented a deep learning based method for multi-modal 3D deformable image registration with structure guidance constraints for adaptive radiotherapy. In our experiments we observed a significant improvement of learning based DIR by incorporation of structure guidance constraints, realized by providing organ segmentations as network input. More precisely, we showed that providing segmentations at first on the reference CT image improves registration results. These segmentations are typically generated and checked by clinical experts during the treatment planning phase and therefore available for all subsequent CT-CBCT registrations. Furthermore, corresponding segmentations on daily CBCT scans become available more easily as learning based segmentation algorithms advance. Incorporation of corresponding segmentations into our deep learning based method yields best results which are comparable to the output of state-of-the-art iterative approaches for structure guided image registration. However, generating deformations over 100 times faster, our learning based approach is capable of application nearly in real-time. Due to its short runtimes and accurate results, our method for structure guided image registration makes adaptive radiotherapy more feasible. It accelerates the clinical workflow and enables a more precise application of radiation doses, so target volumes get irradiated more effectively, while the harm of organs at risk is reduced.
Furthermore, we showed that the ability to build in anatomical knowledge by weakly supervised training of our network improves registration results even when this additional information is not provided during registration of unseen image pairs. Our learning based method does not rely on supervision by hard to obtain ground-truth deformations, but minimizes a suited loss function inspired by variational structure guided registration approaches.
For each registration type, differing in their number of provided inputs, we trained an independent neural network. In future work, we will investigate the implementation of a more flexible approach, handling a variable number of inputs. Additionally, we want to evaluate the integration of supplemental knowledge, especially from segmentations of target volumes that typically do not follow anatomical boundaries.
References
Balakrishnan, G., Zhao, A., Sabuncu, M.R., Guttag, J., Dalca, A.V.: VoxelMorph: a learning framework for deformable medical image registration. IEEE Trans. Med. Imaging 38, 1788–1800 (2019)
Brock, K.K., Mutic, S., McNutt, T.R., Li, H., Kessler, M.L.: Use of image registration and fusion algorithms and techniques in radiotherapy: report of the AAPM radiation therapy committee task group no. 132. Med. Phys. 44(7), e43–e76 (2017)
Elmahdy, M.S., Wolterink, J.M., Sokooti, H., Išgum, I., Staring, M.: Adversarial optimization for joint registration and segmentation in prostate CT radiotherapy. arXiv preprint arXiv:1906.12223 (2019)
Fischer, B., Modersitzki, J.: Curvature based image registration. J. Math. Imaging Vis. 18(1), 81–85 (2003)
Haber, E., Modersitzki, J.: Intensity gradient based registration and fusion of multi-modal images. In: Larsen, R., Nielsen, M., Sporring, J. (eds.) MICCAI 2006. LNCS, vol. 4191, pp. 726–733. Springer, Heidelberg (2006). https://doi.org/10.1007/11866763_89
Han, X.: MR-based synthetic CT generation using a deep convolutional neural network method. Med. Phys. 44(4), 1408–1419 (2017)
Hering, A., Heldmann, S.: Unsupervised learning for large motion thoracic CT follow-up registration. In: Medical Imaging 2019: Image Processing. International Society for Optics and Photonics (2019)
Hering, A., Kuckertz, S., Heldmann, S., Heinrich, M.P.: Enhancing label-driven deep deformable image registration with local distance metrics for state-of-the-art cardiac motion tracking. Bildverarbeitung für die Medizin 2019. I, pp. 309–314. Springer, Wiesbaden (2019). https://doi.org/10.1007/978-3-658-25326-4_69
Himstedt, M., et al.: Deformable image registration using structure guidance for dose accumulation. In: Proceedings of the International Conference on the Use of Computers in Radiation Therapy (ICCR) (2019)
Kuckertz, S., Papenberg, N., Honegger, J., Morgas, T., Haas, B., Heldmann, S.: Deep learning based CT-CBCT image registration for adaptive radio therapy. In: Medical Imaging 2020: Image Processing, vol. 11313, pp. 149–154. International Society for Optics and Photonics, SPIE (2020)
König, L., Rühaak, J., Derksen, A., Lellmann, J.: A matrix-free approach to parallel and memory-efficient deformable image registration. SIAM J. Sci. Comput. 40(3), B858–B888 (2018)
Litjens, G., et al.: A survey on deep learning in medical image analysis. Med. Image Anal. 42, 60–88 (2017)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Rühaak, J., Heldmann, S., Kipshagen, T., Fischer, B.: Highly accurate fast lung CT registration. In: Medical Imaging 2013: Image Processing, vol. 8669, p. 86690Y. International Society for Optics and Photonics (2013)
de Vos, B.D., Berendsen, F.F., Viergever, M.A., Staring, M., Išgum, I.: End-to-end unsupervised deformable image registration with a convolutional neural network. In: Cardoso, M.J., et al. (eds.) DLMIA/ML-CDS-2017. LNCS, vol. 10553, pp. 204–212. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-67558-9_24
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this paper
Cite this paper
Kuckertz, S., Papenberg, N., Honegger, J., Morgas, T., Haas, B., Heldmann, S. (2020). Learning Deformable Image Registration with Structure Guidance Constraints for Adaptive Radiotherapy. In: Špiclin, Ž., McClelland, J., Kybic, J., Goksel, O. (eds) Biomedical Image Registration. WBIR 2020. Lecture Notes in Computer Science(), vol 12120. Springer, Cham. https://doi.org/10.1007/978-3-030-50120-4_5
Download citation
DOI: https://doi.org/10.1007/978-3-030-50120-4_5
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-50119-8
Online ISBN: 978-3-030-50120-4
eBook Packages: Computer ScienceComputer Science (R0)