Abstract
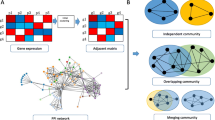
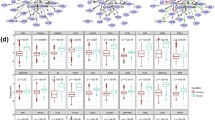
A large amount of applications on high throughput data are applied for cancer diagnosis, clinical treatment and prognosis prediction. Module network inference is an established and effective method to identify the biomarks for specific cancer and uncover the oncogenesis mechanism. Exploiting the overlapping characteristic between modules, rather than detecting disjoint modules, may broaden our understanding of the molecular dysfunction that govern tumor initiation, progression and maintenance. To this end, we propose a novel framework to identify gene modules with overlapping characteristic by integrating gene expression data and protein-protein interaction data. In our framework, social community and overlapping characteristic are introduced to construct disjoint and overlapping modules which can represent the relationship between diverse modules. Applying this framework on six cancer datasets from The Cancer Genome Atlas, we obtain functional gene modules in each cancer, which are more significantly enriched in the known pathways than identified by other state-of-the-art methods. Meanwhile, those identified modules can significantly distinguish the survival prognostic of patients by Kaplan-Meier analysis, which is critical for cancer therapy. Furthermore, identified driver genes in the network can be considered as biomarkers which can distinguish the tumor and normal samples. Uncovering the overlapping feature in gene modules will help elucidate the relationship between modules and could have important therapeutic implication and a more comprehensive interpretation on carcinogenesis.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Lu, X., et al.: DMCM: a data-adaptive mutation clustering method to identify cancer-related mutation clusters. Bioinformatics 35(3), 389–397 (2019)
Hou, J.P., Ma, J.: DawnRank: discovering personalized driver genes in cancer. Genome Med. 6(7), 1–16 (2014)
Gregory, S.: An algorithm to find overlapping community structure in networks. In: Kok, J.N., Koronacki, J., Lopez de Mantaras, R., Matwin, S., Mladenič, D., Skowron, A. (eds.) PKDD 2007. LNCS (LNAI), vol. 4702, pp. 91–102. Springer, Heidelberg (2007). https://doi.org/10.1007/978-3-540-74976-9_12
Zhang, X., et al.: Overlapping community identification approach in online social networks. Phys. A Stat. Mech. Appl. 421, 233–248 (2015)
Yang, J.X., Zhang, X.D.: Finding overlapping communities using seed set. Phys. A Stat. Mech. Appl. 467, 96–106 (2017)
Lee, J., et al.: Improved network community structure improves function prediction. Sci. Rep. 3(2197), 1–9 (2013)
Shendure, J., Ji, H.: Next-generation DNA sequencing. Nat. Biotechnol. 26(10), 1135–1145 (2008)
Hawkins, R.D., et al.: Next-generation genomics: an integrative approach. Nat. Rev. Genet. 11(7), 476–486 (2010)
Tomczak, K., et al.: The cancer genome atlas (TCGA): an immeasurable source of knowledge. Contemp. Oncol. 19(1A), 68–77 (2015)
Ulitsky, I., Shamir, R.: Identification of functional modules using network topology and high-throughput data. BMC Syst. Biol. 1(8), 1–17 (2007)
Ching, T., et al.: Cox-nnet: an artificial neural network method for prognosis prediction of high-throughput omics data. PLoS Comput. Biol. 14(4), e1006076 (2018)
Bonnet, E., et al.: Integrative multi-omics module network inference with lemon-tree. PLoS Comput. Biol. 11(2), e1003983 (2015)
Lu, X., et al.: Driver pattern identification over the gene co-expression of drug response in ovarian cancer by integrating high throughput genomics data. Sci. Rep. 7(1), 16188–16204 (2017)
Lu, X., et al.: The integrative method based on the module-network for identifying driver genes in cancer subtypes. Molecules 23(2), 183–197 (2018)
Ding, Z., et al.: Overlapping community detection based on network decomposition. Sci. Rep. 6(24115), 1–11 (2016)
Wang, Z. et al.: Discovering and profiling overlapping communities in location-based social networks. IEEE Trans. Syst. Man Cybern. Syst. 44(4), 499–509 (2014)
Gui, Q., et al.: A New Method for Overlapping Community Detection Based on Complete Subgraph and Label Propagation, pp. 127–134 (2018)
Amelio, A., Pizzuti, C.: Community mining in signed networks: a multiobjective approach. In: 2013 IEEE/ACM International Conference on Advances in Social Networks Analysis and Mining (ASONAM), pp. 95–99 (2013)
Zhang, J., et al.: Identification of mutated core cancer modules by integrating somatic mutation, copy number variation, and gene expression data. BMC Syst. Biol. 7(S4), 1–12 (2013)
Joshi, A., et al.: Analysis of a Gibbs sampler method for model-based clustering of gene expression data. Bioinformatics 24(2), 176–183 (2008)
Segal, E., et al.: Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat. Genet. 34(4), 166–176 (2003)
Joshi, A., et al.: Module networks revisited: computational assessment and prioritization of model predictions. Bioinformatics 25(4), 490–496 (2009)
Szklarczyk, D., et al.: The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 45, 362–368 (2017). (Database issue)
Gene ontology consortium: the gene ontology (GO) database and informatics resource. Nucleic Acids Res. 32, 258–261 (2004). (Database issue)
Nabavi, S., et al.: EMDomics: a robust and powerful method for the identification of genes differentially expressed between heterogeneous classes. Bioinformatics 32(4), 533–541 (2016)
A Package for Survival Analysis in S. R package version 2.37.7. https://cran.r-project.org/src/contrib/survival_2.44-1.1.tar.gz. Accessed 01 Apr 2019
Heagerty, P.J., Lumley, T., Pepe, M.S.: Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics 56(2), 337–344 (2000)
Lu, X., Peng, X., Deng, Y., Feng, B., Liu, P., Liao, B.: A novel feature selection method based on correlation-based feature selection in cancer recognition. J. Comput. Theor. Nanosci. 11(2), 427–433 (2014)
Burges, C.J.C.: A tutorial on support vector machines for pattern recognition. Data Min. Knowl. Discov. 2(2), 121–167 (1998)
Maere, S., et al.: BiNGO: a cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21(16), 3448–3449 (2005)
Acknowledgement
The authors are grateful to Anagha who provided the initial co-clustering method with GaneSh Java package and Tamaaas who provided the module network construction method with cluster-one Java package. This work was supported by National Natural Science Foundation of China (Grant Nos. 61502159, 61472467 and 61672011), Natural Science Foundation of Hunan Province, China (Grant No. 2018JJ2053) and National Key R&D Program of China (2017YFC1311003).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Lu, X. et al. (2019). The Detection of Gene Modules with Overlapping Characteristic via Integrating Multi-omics Data in Six Cancers. In: Huang, DS., Jo, KH., Huang, ZK. (eds) Intelligent Computing Theories and Application. ICIC 2019. Lecture Notes in Computer Science(), vol 11644. Springer, Cham. https://doi.org/10.1007/978-3-030-26969-2_38
Download citation
DOI: https://doi.org/10.1007/978-3-030-26969-2_38
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-26968-5
Online ISBN: 978-3-030-26969-2
eBook Packages: Computer ScienceComputer Science (R0)