Abstract
We propose a method for automatic segmentation of the prostate clinical target volume for brachytherapy in transrectal ultrasound (TRUS) images. Because of the large variability in the strength of image landmarks and characteristics of artifacts in TRUS images, existing methods achieve a poor worst-case performance, especially at the prostate base and apex. We aim at devising a method that produces accurate segmentations on easy and difficult images alike. Our method is based on a novel convolutional neural network (CNN) architecture. We propose two strategies for improving the segmentation accuracy on difficult images. First, we cluster the training images using a sparse subspace clustering method based on features learned with a convolutional autoencoder. Using this clustering, we suggest an adaptive sampling strategy that drives the training process to give more attention to images that are difficult to segment. Secondly, we train multiple CNN models using subsets of the training data. The disagreement within this CNN ensemble is used to estimate the segmentation uncertainty due to a lack of reliable landmarks. We employ a statistical shape model to improve the uncertain segmentations produced by the CNN ensemble. On test images from 225 subjects, our method achieves a Hausdorff distance of \(2.7\,\pm \,2.1\) mm, Dice score of \(93.9\,\pm \,3.5\), and it significantly reduces the likelihood of committing large segmentation errors.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
1 Introduction
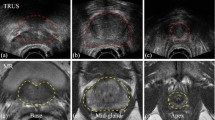
Transrectal ultrasound (TRUS) is routinely used in the diagnosis and treatment of prostate cancer. This study addresses the segmentation of the clinical target volume (CTV) in 2D TRUS images, an essential step for radiation treatment planning [9]. The CTV is delineated on a series of 2D TRUS images from the prostate base to apex. This is a challenging task because image landmarks are often weak or non-existent, especially at the base and apex, and various types of artifacts can be present. Therefore, manual segmentation is tedious and prone to high inter-observer variability. Several semi- and fully-automatic segmentation algorithms have been proposed based on methods such as level sets, shape and appearance models, and machine learning [7, 10]. However, these methods often require careful initialization and are too slow for real-time segmentation. Moreover, although some of them achieve good average results in terms of, e.g., Dice Similarity Coefficient (DSC), criteria that show worst-case performance such as the Hausdorff Distance (HD) are either not reported or display large variances. This is because some images can be particularly difficult to segment due to weak prostate edges and strong artifacts. This also poses a challenge for deep learning-based methods that have achieved great success in medical image segmentation. Since they have a high representational power and are trained using stochastic gradient descent with uniform sampling of the training data, their training can be dominated by the more typical samples in the training set, leading to poor generalization on less-represented images.
In this paper, we propose a method for segmentation of the CTV in 2D TRUS images that is geared towards achieving good results on most test images while at the same time reducing large segmentation errors. Our contributions are:
-
1.
We propose a novel convolutional neural network (CNN) architecture for segmentation of the CTV in 2D TRUS images.
-
2.
We suggest an adaptive sampling method for CNN training. In brief, our method samples the training images based on how likely they are to contribute to improving the segmentation of difficult images in a validation set.
-
3.
We estimate the segmentation uncertainty based on the disagreement among an ensemble of CNNs and propose a novel method to improve the highly uncertain segmentations with the help of a statistical shape model (SSM).
2 Materials and Methods
2.1 Data
We used the TRUS images of 675 subjects. From each subject, 7 to 14 2D TRUS images of size \(415 \times 490\) pixels with a pixel size of \(0.15\,\times \,0.15\) mm\(^2\) were acquired. The CTV was delineated in each slice by experienced radiation oncologists. We used the data from 450 subjects for training, including cross-validation, and left the remaining 225 subjects (including a total of 2207 2D images) for test.
2.2 Clustering of the Training Images
We rely on the method of sparse subspace clustering [1] and use a convolutional autoencoder (CAE) for learning low-dimensional image representations as proposed in [4]. As shown in Fig. 1, the encoder part of the CAE learns a low-dimensional representation \(z^i_{\text {enc}}\) for an input image \(x^i\). Then, a fully-connected layer, which consists of multiplication with a matrix, \(\varGamma \), without a bias term and nonlinear activation function, transforms this representation into the input to the decoder, \(z^i_{\text {dec}}\). The sparse subspace clustering is enforced by requiring:

where \(Z_{\text {enc}}\) is the matrix that has \(z^i_{\text {enc}}\) for all training images as its columns, and similarly for \(Z_{\text {dec}}\), and \(\varGamma \) is a sparse matrix with zero diagonal. By enforcing sparsity on \(\varGamma \), we require that the representation of the \(i^{\text {th}}\) image, \(z^i_{\text {dec}}\), be approximated as a linear combination of a small number of those of other images in the training set. Note that although the relation between \(Z_{\text {dec}}\) and \(Z_{\text {enc}}\) is linear, the clustering method is far from linear because \(z^i_{\text {enc}}\) is a very rich and highly non-linear representation of the image.
We first train a standard CAE, i.e., with \(\varGamma = I\). In this stage, we minimize the standard CAE cost function, i.e., the reconstruction error \(\Vert \hat{X}- X\Vert ^2_2\), where X and \(\hat{X}\) denote, respectively, matrices of the input images and the reconstructed images. In the second stage, we introduce \(\varGamma \) and train the network by solving:

We empirically chose \(\lambda _1= \lambda _2= 0.1\). For both training stages, we trained the network for 100 epochs using Adam [5] with a learning rate of \(10^{-3}\). Once the network is trained, an affinity matrix can be created as \(C= | \varGamma | + | \varGamma ^{\text {T}} |\), where C(i, j) indicates the similarity between the \(i^{\text {th}}\) and \(j^{\text {th}}\) images. Spectral clustering methods can be used to cluster the data based on C, but we will use C directly as explained in Sect. 2.4.
2.3 Proposed CNN Architecture
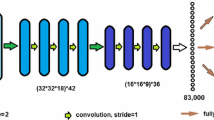
A simplified representation of our CNN is shown in Fig. 2. Our design is different from widely-used networks such as [8] in that: (1) We apply convolutional filters of varying sizes (\(k \in \{3, 5, 7, 9, 11\}\)) and strides (\(s \in \{1, 2, 3, 4, 5\}\)) directly to the input image to extract fine and coarse features. Because small image patches are overwhelmed by speckle and contain little edge information, applying larger filters directly on the image should help the network learn more informative features at different scales, (2) The computed features at each fine scale are forwarded to all coarser layers by applying convolutional kernels of proper sizes and strides. This promotes feature reuse, which reduces the number of network parameters while increasing the richness of the learned representations [3]. Hence, the network extracts features at multiple different resolutions and fields-of-view. These features are then combined via a series of transpose convolutions. (3) In both the contracting and the expanding paths, features go through residual blocks to increase the richness of representations and ease the training. The network outputs a prostate segmentation probability map (in [0,1]). We train the network by maximizing the DSC between this probability map and the ground-truth segmentation. For this, we used Adam with a learning rate of \(10^{-4}\) and performed 200 epochs. The training process is explained in Sect. 2.4.
2.4 Training a CNN Ensemble with Adaptive Sampling
Due to non-convexity and extreme complexity of their optimization landscape, deep CNNs converge to a local minimum. With small training data, these minima can be heavily influenced by the more prevalent samples in the training set. A powerful approach to reducing the sensitivity to local minima and reducing the generalization error is to learn an ensemble of models [2]. We train \(K= 5\) CNN models using 5-fold cross validation. Let us denote the indices of the training and validation images for one of these models with \(S_{\text {tr}}\) and \(S_{\text {vl}}\), respectively. Let \(e_i\) denote the “error" committed on the \(i^{\text {th}}\) validation image by the CNN after the current training epoch. As shown in Fig. 3, for the next epoch we sample the training images according to their similarity to the difficult validation images. Specifically, we compute the probability of sampling the \(j^{\text {th}}\) training image as:

We initialize p to a uniform distribution for the first epoch. Importantly, there is a great flexibility in the choice of the error, e. For example, e does not have to possess requirements such as differentiability. In this work, we chose the Hausdorff Distance (HD) as e. For two curves, X and Y, HD is defined as  . Although HD is an important measure of segmentation error, it cannot be easily minimized as it is non-differentiable. Our approach provides an indirect way to reduce HD.
. Although HD is an important measure of segmentation error, it cannot be easily minimized as it is non-differentiable. Our approach provides an indirect way to reduce HD.
Top: an “easy” image, (a) the CNNs produce similar results, (b) the final segmentation produced by thresholding the mean probability map. Bottom: a “difficult” image, (c) there is large disagreement between CNNs, (d) the certainty map with \(s_{\text {init}}\) (red) superimposed, (e) the final segmentation, \(s_{\text {impr}}\) (blue), obtained using SSM.
2.5 Improving Uncertain Segmentations Using an SSM
Training multiple models enables us to estimate the segmentation (un)certainty by examining the disagreement among the models. For a given image, we compute the average pair-wise DSC between the segmentations produced by the 5 CNNs. If this value is above the empirically-chosen threshold of 0.95, we trust the CNN segmentations because of high agreement among the 5 CNNs trained on different data. In such a case, we will compute the average of the 5 probability maps and threshold it at 0.50 to yield the final segmentation (Fig. 4, top row).
If the mean pair-wise DSC among the 5 CNN segmentations is below 0.95, we improve it by introducing prior information in the form of an SSM. We built the SSM from a set of 75 MR images with ground-truth prostate segmentation provided by expert radiologists. From each slice of the MR images, we extracted 100 equally-spaced points on the boundary of the prostate, rigidly (i.e., translation, scale, and rotation) registered them to one reference point set, and computed the SVD of the point sets. We built three separate SSMs for base, mid-gland, and apex. In deciding whether an MRI slice belonged to base, mid-gland, or apex, we assumed that each of these three sections accounted for one third of the prostate length. We use u and V to denote, respectively, the mean shape and the matrix with the n most important shape modes as its columns. We chose \(n=5\) because the top 5 modes explained more than \(98 \%\) of the shape variance.
If the agreement among the CNN segmentations is below the threshold, we use them to compute: (1) An initial segmentation boundary, \(s_{\text {init}}\), by thresholding the average of the 5 probability maps, \(\bar{p}\), at 0.5, and (2) a certainty map:
where \(F_{\text {KW}}\) is based on the Kohavi-Wolpert variance [6]. \(F_{\text {KW}}\) is 0 where all models agree and increases as the disagreement grows. As shown in Fig. 4(d), Q indicates, roughly, the locations where segmentation boundaries predicted by the 5 models are close, i.e., segmentations are more likely to be correct. Therefore, we estimate an improved segmentation boundary, \(s_{\text {impr}}\), as:
where t, s, and w denote, respectively, translation, scale, and the coefficients of the shape model, \(R_{\theta }\) is the rotation matrix with angle \(\theta \), and \(\Vert . \Vert _Q \) denotes the weighted \(\ell _2\) norm using weights Q computed in Eq. (4). In other words, we fit an SSM to \(s_{\text {init}}\) while attaching more importance to parts of \(s_{\text {init}}\) that have higher certainty. Since the objective function in Eq. 5 is non-convex, alternating minimization is used to find a stationary point. We initialize t to the centroid of the initial segmentation, s to 1, and w and \(\theta \) to zero and perform alternating minimization until the objective function reduces by less than \(1 \%\) in an iteration. Up to 3 iterations sufficed to converge to a good result (Fig. 4, bottom row).
3 Results and Discussion
We compare our method with the adaptive shape model-based method of [10] and CNN model of [8], which we denote as ADSM and U-NET, respectively. We report three results for our method: (1) Proposed-OneCNN: only one CNN is trained, (2) Proposed-Ensemble: five CNNs are trained as explained in Sect. 2.4 and the final segmentation is obtained by thresholding the average probability map at 0.5, and (3) Proposed-Full: improves uncertain segmentations produced by Proposed-Ensemble as explained in Sect. 2.5. Our comparison criteria are DSC and HD. We also report the \(95 \%\)-percentile of HD across the test images as a measure of the worst-case performance on the population of test images.
As shown in Table 1, our method outperformed the other methods in terms of DSC and HD. Paired t-tests (at \(p=0.01\)) showed that the HD obtained by our method was significantly smaller than the other methods in all three prostate sections. Our method also achieved much smaller values for the \(95 \%\)-percentile of HD. Figure 5 shows example segmentations produced by different methods.
Table 2 shows the effectiveness of our proposed strategies for improving the segmentations. Proposed-Ensemble and Proposed-Full achieve much better results than Proposed-OneCNN. There is a marked improvement in DSC. The reduction in HD is also substantial. Mean, standard deviation, and the \(95 \%\)-percentile of HD have been greatly reduced by our proposed strategies. Paired t-tests (at \(p=0.01\)) showed that Proposed-Ensemble achieved a significantly lower HD than Proposed-OneCNN and, on images that were processed by SSM fitting, Proposed-Full significantly reduced HD compared with Proposed-Ensemble.
Both the CAE (Fig. 1) and the CNN (Fig. 2) were implemented in TensorFlow. On an Nvidia GeForce GTX TITAN X GPU, the training times for the CAE and each of the CNNs, respectively, were approximately 24 and 12 h. For a test image, each CNN produces a segmentation in 0.02 s.
4 Conclusion
In the context of prostate CTV segmentation in TRUS, we proposed adaptive sampling of the training data, ensemble learning, and use of prior shape information to improve the segmentation accuracy and robustness and reduce the likelihood of committing large segmentation errors. Our method achieved significantly better results than competing methods in terms of HD, which measures largest segmentation error. Our methods also substantially reduced the maximum errors committed on the population of test images. An important contribution of this work was a method to compute a segmentation certainty map, which we used to improve the segmentation accuracy with the help of an SSM. This certainty map can have many other useful applications, such as in registration of TRUS to pre-operative MRI and for radiation treatment planning. A shortcoming of this work is with regard to our ground-truth segmentations, which have been provided by expert radiation oncologists on TRUS images. These segmentations can be biased at the prostate base and apex. Therefore, a comparison with registered MRI is warranted.
References
Elhamifar, E., Vidal, R.: Sparse subspace clustering: algorithm, theory, and applications. IEEE Trans. Pattern Anal. Mach. Intell. 35(11), 2765–2781 (2013)
Goodfellow, I., Bengio, Y., Courville, A., Bengio, Y.: Deep Learning, vol. 1. MIT Press, Cambridge (2016)
Huang, G., Liu, Z., Weinberger, K.Q., van der Maaten, L.: Densely connected convolutional networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, vol. 1, p. 3 (2017)
Ji, P., Zhang, T., Li, H., Salzmann, M., Reid, I.: Deep subspace clustering networks. In: Advances in Neural Information Processing Systems, pp. 23–32 (2017)
Kingma, D.P., Ba, J.: Adam: a method for stochastic optimization. In: Proceedings of the 3rd International Conference on Learning Representations (ICLR) (2014)
Kuncheva, L., Whitaker, C.: Measures of diversity in classifier ensembles and their relationship with the ensemble accuracy. Mach. Learn. 51(2), 181–207 (2003)
Qiu, W., Yuan, J., Ukwatta, E., Fenster, A.: Rotationally resliced 3D prostate trus segmentation using convex optimization with shape priors. Med. phys. 42(2), 877–891 (2015)
Ronneberger, O., Fischer, P., Brox, T.: U-net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Sylvester, J.E., Grimm, P.D., Eulau, S.M., Takamiya, R.K., Naidoo, D.: Permanent prostate brachytherapy preplanned technique: the modern seattle method step-by-step and dosimetric outcomes. Brachytherapy 8(2), 197–206 (2009)
Yan, P., Xu, S., Turkbey, B., Kruecker, J.: Adaptively learning local shape statistics for prostate segmentation in ultrasound. IEEE Trans. Biomed. Eng. 58(3), 633–641 (2011)
Acknowledgment
This work was supported by Prostate Cancer Canada, the CIHR, the NSERC, and the C.A. Laszlo Chair in Biomedical Engineering held by S. Salcudean.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
Karimi, D. et al. (2018). Accurate and Robust Segmentation of the Clinical Target Volume for Prostate Brachytherapy. In: Frangi, A., Schnabel, J., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds) Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. MICCAI 2018. Lecture Notes in Computer Science(), vol 11073. Springer, Cham. https://doi.org/10.1007/978-3-030-00937-3_61
Download citation
DOI: https://doi.org/10.1007/978-3-030-00937-3_61
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-00936-6
Online ISBN: 978-3-030-00937-3
eBook Packages: Computer ScienceComputer Science (R0)