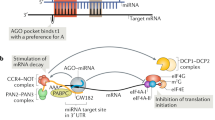
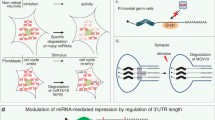
Abstract
Trim-NHL proteins are defined by RING, B-Box and Coiled-coil protein motifs (referred to collectively as the Trim domain) coupled to an NHL domain. The C. elegans, D. melanogaster, mouse and human Trim-NHL proteins are potential and in several cases confirmed, E3 ubiquitin ligases. Current research is focused on identifying targets and pathways for Trim-NHL-mediated ubiquitination and in assessing the contribution of the NHL protein-protein interaction domain for function and specificity. Several Trim-NHL proteins were discovered in screens for developmental genes in model organisms; mutations in one of the family members, Trim32, cause developmental disturbances in humans. In most instances, mutations that alter protein function map to the NHL domain. The NHL domain is a scaffold for the assembly of a translational repressor complex by the Brat proto-oncogene, a well-studied family member in Drosophila. The link to translational control is common to at least four Trim-NHLs that associate with miRNA pathway proteins. So far, two have been shown to repress (Mei-P26 and Lin41) and two to promote (NHL-2, Trim32) miRNA-mediated gene silencing. In this chapter we will describe structure-function relations for each of the proteins and then focus on the lessons being learned from these proteins about miRNA functions in development and in stem cell biology.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Reinhart BJ, Slack FJ, Basson M et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000; 403(6772):901–906.
Pasquinelli AE, Reinhart BJ, Slack F et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 2000; 408(6808):86–89.
Lee R, Feinbaum R, Ambros V. A short history of a short RNA. Cell 2004; 116(2 Suppl):S89–92, 81.
Slack FJ, Basson M, Liu Z et al. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell 2000; 5(4):659–669.
Slack FJ, Ruvkun G. A novel repeat domain that is often associated with RING finger and B-box motifs. Trends Biochem Sci 1998; 23(12):474–475.
Meroni G, Diez-Roux G. TRIM/RBCC, a novel class of’ single protein RING finger’ E3 ubiquitin ligases. Bioessays 2005; 27(11): 1147–1157.
Sardiello M, Cairo S, Fontaneila B et al. Genomic analysis of the TRIM family reveals two groups of genes with distinct evolutionary properties. BMC Evol Biol 2008; 8:225.
Nisole S, Stoye JP, Saib A. TRIM family proteins: retroviral restriction and antiviral defence. Nat Rev Microbiol 2005; 3(10):799–808.
Reymond A, Meroni G, Fantozzi A et al. The tripartite motif family identifies cell compartments. EMBO J 2001;20(9):2140–2151.
Hyenne V, Desrosiers M, Labbe JC. C. elegans Brat homologs regulate PAR protein-dependent polarity and asymmetric cell division. Dev Biol 2008; 321(2):368–378.
O’Farrell F, Esfahani SS, Engstrom Y et al. Regulation of the Drosophila lin-41 homologue dappledbylet-7reveals conservation of a regulatory mechanism within the LIN-41 subclade. Dev Dyn 2008; 237(1): 196–208.
Saurin AJ, Borden KL, Boddy MN et al. Does this have a familiar RING? Trends Biochem Sci 1996; 21(6):208–214.
Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem 2009; 78:399–434.
Joazeiro CA, Wing SS, Huang H et al. The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science 1999;286(5438):309–312.
Yokouchi M, Kondo T, Houghton A et al. Ligand-inducedubiquitination of the epidermal growth factorreceptor involves the interaction of the c-Cbl RING finger and UbcH7. J Biol Chem 1999; 274(44):31707–31712.
Lorick KL, Jensen JP, Fang S et al. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci USA 1999; 96(20):11364–11369.
Freemont PS. RING for destruction? Curr Biol 2000; 10(2):R84–87.
Lupas A. Coiled coils: new structures and new functions. Trends Biochem Sci 1996; 21(10):375–382.
Zheng N, Wang P, Jeffrey PD et al. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell 2000; 102(4):533–539.
Horn EJ, Albor A, Liu Y et al. RING protein Trim32 associated with skin carcinogenesis has anti-apoptotic and E3-ubiquitin ligase properties. Carcinogenesis 2004; 25(2): 157–167.
Kudryashova E, Kudryashov D, Kramerova I et al. Trim32 is a ubiquitin ligase mutated in limb girdle muscular dystrophy type 2H that binds to skeletal muscle myosin and ubiquitinates actin. J Mol Biol 2005; 354(2):413–424.
Balastik M, Ferraguti F, Pires-da Silva A et al. Deficiency in ubiquitin ligase TRIM2 causes accumulation of neurofilament light chain and neurodegeneration. Proc Nat1 Acad Sci USA 2008; 105(33): 12016–12021.
Rybak A, Fuchs H, Hadian K et al. The let-7 target gene mouse lin-41 is a stem cell specific E3 ubiquitin ligase for the miRNA pathway protein Ago2. Nat Cell Biol 2009; 11(12):1411–1420.
Chiang AP, Beck JS, Yen HJ et al. Homozygosity mapping with SNP arrays identifies TRIM32, an E3 ubiquitin ligase, as a Bardet-Biedl syndrome gene (BBS 11). Proc Natl Acad Sci USA 2006; 103(16):6287–6292.
Saccone V, Palmieri M, Passamano L et al. Mutations that impair interaction properties of TRIM32 associated with limb-girdle muscular dystrophy 2H. Hum Mutat 2008; 29(2):240–247.
Massiah MA, Matts JA, Short KM et al. Solution structure ofthe MID lB-box2CHC(D/C)C(2)H(2) zinc-binding domain: insights into an evolutionarily conserved RING fold. J Mol Biol 2007; 369(1): 1–10.
Massiah MA, Simmons BN, Short KM et al. Solution structure of the RBCC/TRIM B-boxl domain of human MIDI: B-box with a RING. J Mol Biol 2006; 358(2):532–545.
Tao H, Simmons BN, Singireddy S et al. Structure of the MIDI tandem B-boxes reveals an interaction reminiscent of intermolecular ring heterodimers. Biochemistry 2008; 47(8):2450–2457.
Grigoryan G, Keating AE. Structural specificity in coiled-coil interactions. Curr Opin Struct Biol 2008; 18(4):477–483.
Zhou AX, Hartwig JH, Akyurek LM. Filamins in cell signaling, transcription and organ development. Trends Cell Biol.
Arama E, Dickman D, Kimchie Z et al. Mutations in the beta-propeller domain of the Drosophila brain tumor (brat) protein induce neoplasm in the larval brain. Oncogene 2000; 19(33):3706–3716.
Sonoda J, Wharton RP. Drosophila brain tumor is a translational repressor. Genes Dev 2001; 15(6):762–773.
Edwards TA, Wilkinson BD, Wharton RP et al. Model of the brain tumor-Pumilio translation repressor complex. Genes Dev 2003; 17(20):2508–2513.
Pickart CM, Cohen RE. Proteasomes and their kin: proteases in the machine age. Nat Rev Mol Cell Biol 2004; 5(3): 177–187.
Schulman BA, Harper JW. Ubiquitin-like proteinactivation by El enzymes: the apex for downstream signalling pathways. Nat Rev Mol Cell Biol 2009; 10(5):319–331.
Albor A, El-Hizawi S, Horn EJ et al. The interaction of Piasy with Trim32, an E3-ubiquitin ligase mutated in limb-girdlemusculardystrophytype 2H, promotes Piasy degradation andregulatesUVB-inducedkeratinocyte apoptosis through NFkappaB. J Biol Chem 2006; 281(35):25850–25866.
Schwamborn JC, Berezikov E, Knoblich JA The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell 2009; 136(5):913–925.
Locke M, Tinsley CL, Benson MA et al. TRIM32 is an E3 ubiquitin ligase for dysbindin. Hum Mol Genet 2009; 18(13):2344–2358.
Fridell RA, Harding LS, Bogerd HP et al. Identification of a novel human zinc finger protein that specifically interacts with the activation domain of lentiviral Tat proteins. Virology 1995; 209(2):347–357.
Frosk P, Weiler T, Nylen E et al. Limb-girdle muscular dystrophy type 2H associated withmutationin TRIM32, a putative E3-ubiquitin-ligase gene. Am J Hum Genet 2002; 70(3):663–672.
Li W, Zhang Q, Oiso N et al. Hermansky-Pudlak syndrome type 7 (HPS-7) results from mutant dysbindin, a member of the biogenesis of lysosome-related organelles complex 1 (BLOC-1). Nat Genet 2003; 35(l):84–89.
Mykytyn K, Sheffield VC. Establishing a connection between cilia and Bardet-Biedl Syndrome. Trends Mol Med 2004; 10(3):106–109.
Ansley SJ, Badano JL, Blacque OE et al. Basal body dysfunction is a likely cause of pleiotropic Bardet-Biedl syndrome. Nature 2003; 425(6958):628–633.
Kudryashova E, Wu J, Havton LA et al. Deficiency of the E3 ubiquitin ligase TRIM32 in mice leads to a myopathy with a neurogenic component. Hum Mol Genet 2009; 18(7): 1353–1367.
Pasquinelli AE, Ruvkun G. Control of developmental timing by micrornas and their targets. Annu Rev Cell Dev Biol 2002; 18:495–513.
Kloosterman WP, Wienholds E, Ketting RF et al. Substrate requirements for let-7 function in the developing zebrafish embryo. Nucleic Acids Res 2004; 32(21):6284–6291.
Lancman JJ, Caruccio NC, Harfe BD et al. Analysis of the regulation of lin-41 during chick and mouse limb development. Dev Dyn 2005; 234(4):948–960.
Schulman BR, Esquela-Kerscher A, Slack FJ. Reciprocal expression of lin-41 and the microRNAs let-7 and mir-125 during mouse embryogenesis. Dev Dyn 2005; 234(4): 1046–1054.
Kanamoto T, Terada K, Yoshikawa H et al. Cloning and regulation of the vertebrate homologue of lin-41 that functions as a heterochronic gene in Caenorhabditis elegans. Dev Dyn 2006; 235(4): 1142–1149.
Rodriguez A, Zhou Z, Tang ML et al. Identification of immune system and response genes and novel mutations causing melanotic tumor formation in Drosophila melanogaster. Genetics 1996; 143(2):929–940.
Brody T, Stivers C, Nagle J et al. Identification of novel Drosophila neural precursor genes using a differential embryonic head cDNA screen. Mech Dev 2002; 113(1):41–59.
O’Farrell F, Kylsten P. A mis-expression study of factors affecting Drosophila PNS cell identity. Biochem Biophys Res Commun 2008; 370(4):657–662.
Loer B, Bauer R, Bornheim R et al. The NHL-domain protein Wech is crucial for the integrin-cytoskeleton link. Nat Cell Biol 2008; 10(4):422–428.
Delon I, Brown N. Cell-matrix adhesion: the wech connection. Curr Biol 2008; 18(9):R389–391.
Loer B, Hoch M. Wech proteins: roles in integrin functions and beyond. Cell Adh Migr 2008; 2(3):177–179.
Sokol NS, Xu P, Jan YN et al. Drosophila let-7 microRNA is required for remodeling of the neuromusculature during metamorphosis. Genes Dev 2008; 22(12):1591–1596.
El Husseini AE, Vincent SR Cloning and characterization of a novel RING finger protein that interacts with class V myosins. J Biol Chem 1999; 274(28):19771–19777.
Trybus KM. Myosin V from head to tail. Cell Mol Life Sci 2008; 65(9): 1378–1389.
Yan Q, Sun W, Kujala P et al. CART: an Hrs/actinin-4/BERP/myosin V protein complex required for efficient receptor recycling. Mol Biol Cell 2005; 16(5):2470–2482.
Mosesson Y, Chetrit D, Schley L et al. Monoubiquitinylation regulates endosomal localization of Lst2, a negative regulator of EGF receptor signaling. Dev Cell 2009; 16(5):687–698.
Zwang Y, Yarden Y. Systems biology of growth factor-induced receptor endocytosis. Traffic 2009; 10(4):349–363.
Ohkawa N, Kokura K, Matsu-Ura T et al. Molecular cloning and characterization of neural activity-related RING finger protein (NARF): a new member of the RBCC family is a candidate for the partner of myosin V. J Neurochem 2001; 78(1):75–87.
Gray PA, Fu H, Luo P et al. Mouse brain organization revealed through direct genome-scale TF expression analysis. Science 2004; 306(5705):2255–2257.
Wright TR. The Wilhelmine E. Key 1992 Invitational lecture. Phenotypic analysis of the Dopa decarboxylase gene cluster mutants in Drosophila melanogaster. J Hered 1996; 87(3):175–190.
Neumuller RA, Betschinger J, Fischer A et al. Mei-P26 regulates microRNAs and cell growth in the Drosophila ovarian stem cell lineage. Nature 2008; 454(7201):241–245.
Frank DJ, Edgar BA, Roth MB. The Drosophila melanogaster gene brain tumor negatively regulates cell growth and ribosomal RNA synthesis. Development 2002; 129(2):399–407.
Bello B, Reichert H, Hirth F. The brain tumor gene negatively regulates neural progenitor cell proliferation in the larval central brain of Drosophila. Development 2006; 133(14):2639–2648.
Bowman SK, Rolland V, Betschinger J et al. The tumor suppressors Brat and Numb regulate transit-amplifying neuroblast lineages in Drosophila. Dev Cell 2008; 14(4):535–546.
Knoblich JA. Mechanisms of asymmetric stem cell division. Cell 2008; 132(4):583–597.
Kohlmaier A, Edgar BA. Proliferative control in Drosophila stem cells. Curr Opin Cell Biol 2008; 20(6):699–706.
Betschinger J, Mechtler K, Knoblich JA. Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell 2006; 124(6):1241–1253.
Lee CY, Wilkinson BD, Siegrist SE et al. Brat is a Miranda cargo protein that promotes neuronal differentiation and inhibits neuroblast self-renewal. Dev Cell 2006; 10(4):441–449.
Sekelsky JJ, McKim KS, Messina L et al. Identification of novel Drosophila meiotic genes recovered in a P-element screen. Genetics 1999; 152(2):529–542.
Page SL, McKim KS, Deneen B et al. Genetic studies of mei-P26 reveal a link between the processes that control germ cell proliferation in both sexes and those that control meiotic exchange in Drosophila. Genetics 2000; 155(4):1757–1772.
Ivanov AI, Rovescalli AC, Pozzi P et al. Genes required for Drosophila nervous system development identified by RNA interference. Proc Natl Acad Sci USA 2004; 101(46):16216–16221.
Glasscock E, Singhania A, Tanouye MA. The mei-P26 gene encodes a RING finger B-box coiled-coil-NHL protein that regulates seizure susceptibility in Drosophilia. Genetics 2005; 170(4):1677–1689.
Pavlidis P, Ramaswami M, Tanouye MA. The Drosophila easily shocked gene: a mutation in a phospholipid synthetic pathway causes seizure, neuronal failure and paralysis. Cell 1994; 79(1):23–33.
Pascual A, Chaminade M, Preat T. Ethanolamine kinase controls neuroblast divisions in Drosophila mushroom bodies. Dev Biol 2005; 280(1): 177–186.
Okamura K, Ishizuka A, Siomi H et al. Distinct roles for argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev 2004; 18(14):1655–1666.
Park JK, Liu X, Strauss TJ et al. The miRNA pathway intrinsically controls self-renewal of Drosophila germline stem cells. Curr Biol 2007; 17(6):533–538.
Jin Z, Xie T. Dcr-1 maintains Drosophila ovarian stem cells. Curr Biol 2007; 17(6):539–544.
Hammell CM, Lubin I, Boag PR et al. nhl-2 Modulates microRNA activity in Caenorhabditis elegans. Cell 2009; 136(5):926–938.
Chu CY, Rana TM. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol 2006; 4(7):e210.
Loedige I, Filipowicz W. TRIM-NHL proteins take on miRNA regulation. Cell 2009; 136(5):818–820.
Coller JM, Tucker M, Sheth U et al. The DEAD box helicase, Dhhlp, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA 2001; 7(12):1717–1727.
Eulalio A, Rehwinkel J, Stricker M et al. Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing. Genes Dev 2007; 21(20):2558–2570.
Farkas LM, Huttner WB. The cell biology of neural stem and progenitor cells and its significance for their proliferation versus differentiation during mammalian brain development. Curr Opin Cell Biol 2008; 20(6):707–715.
Zhong W, Chia W. Neurogenesis and asymmetric cell division. Curr Opin Neurobiol 2008; 18(1):4–11.
Kosodo Y, Huttner WB. Basal process and cell divisions of neural progenitors in the developing brain. Dev Growth Differ 2009; 51(3):251–261.
Bussing I, Slack FJ, Grosshans H. let-7 microRNAs in development, stem cells and cancer. Trends Mol Med 2008; 14(9):400–409.
Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature 2010.
Knoepfler PS, Cheng PF, Eisenman RN. N-mycis essential duringneurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation. Genes Dev 2002; 16(20):2699–2712.
Martins RA, Zindy F, Donovan S et al. N-myc coordinates retinal growth with eye size during mouse development. Genes Dev 2008; 22(2): 179–193.
Kloosterman WP, Wienholds E, de Bruijn E et al. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat Methods 2006; 3(1):27–29.
Nishino J, Kim I, Chada K et al. Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing pl6Ink4a and pl9 Arf Expression. Cell 2008; 135(2):227–239.
Rybak A, Fuchs H, Smirnova L et al. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol 2008; 10(8):987–993.
Mailer Schulman BR, Liang X, Stahlhut C et al. The let-7 microRNA target gene, Mlin41/Trim71 is required for mouse embryonic survival and neural tube closure. Cell Cycle 2008; 7(24):3935–3942.
Duchaine TF, Wohlschlegel JA, Kennedy S et al. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell 2006; 124(2):343–354.
Diederichs S, Haber DA. Dual role for argonautes in microRNA processing andpost-transcriptional regulation of microRNA expression. Cell 2007; 131(6):1097–1108.
Chatterjee S, Grosshans H. Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature 2009; 461(7263):546–549.
Sinkkonen L, Hugenschmidt T, Berninger P et al. microRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stemcells. Nat Struct Mol Biol 2008;15(3):259–267.
Dueck A, Meister G. TRIMming microRNA function in mouse stem cells. Nat Cell Biol 2009; 11(12):1392–1393.
Selbach M, Schwanhausser B, Thierfelder N et al. Widespread changes in protein synthesis induced by microRNAs. Nature 2008; 455(7209):58–63.
Ding XC, Slack FJ, Grosshans H. The let-7 microRNA interfaces extensively with the translation machinery to regulate cell differentiation. Cell Cycle 2008; 7(19):3083–3090.
Tokumaru S, Suzuki M, Yamada H et al. let-7 regulates Dicer expression and constitutes a negative feedback loop. Carcinogenesis 2008.
Johnson CD, Esquela-Kerscher A, Stefani G et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res 2007; 67(16):7713–7722.
Ashraf SI, McLoon AL, Sclarsic SM et al. Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell 2006; 124(1):191–205.
Gibbings DJ, Ciaudo C, Erhardt M et al. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol 2009; 11(9):1143–1149.
Kotaja N, Bhattacharyya SN, Jaskiewicz L et al. The chromatoid body of male germ cells: Similarity with processing bodies and presence of Dicer and microRNA pathway components. Proc Natl Acad Sci USA 2006.
Wulczyn FG, Smirnova L, Rybak A et al. Post-transcriptional regulation of the let-7 microRNA during neural cell specification. FASEB J 2007; 21(2):415–426.
Lee YS, Pressman S, Andress AP et al. Silencing by small RNAs is linked to endosomal trafficking. Nat Cell Biol 2009; 11(9):1150–1156.
Kirilly D, Xie T. The Drosophila ovary: an active stem cell community. Cell Res 2007; 17(1): 15–25.
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2010 Landes Bioscience and Springer Science+Business Media, LLC
About this chapter
Cite this chapter
Wulczyn, F.G., Cuevas, E., Franzoni, E., Rybak, A. (2010). miRNAs Need a Trim. In: Großhans, H. (eds) Regulation of microRNAs. Advances in Experimental Medicine and Biology, vol 700. Springer, New York, NY. https://doi.org/10.1007/978-1-4419-7823-3_9
Download citation
DOI: https://doi.org/10.1007/978-1-4419-7823-3_9
Publisher Name: Springer, New York, NY
Print ISBN: 978-1-4419-7822-6
Online ISBN: 978-1-4419-7823-3
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)