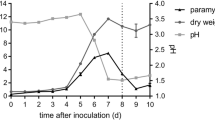
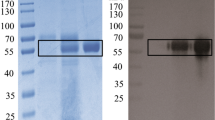
Abstract
Prokaryotes are known to produce and secrete a broad range of biopolymers with a high functional and structural heterogeneity, often with critical duties in the bacterial physiology and ecology. Among these, exopolysaccharides (EPS) play relevant roles in the interaction of bacteria with eukaryotic hosts. EPS can help to colonize the host and assist in bacterial survival, making this interaction more robust by facilitating the formation of structured biofilms. In addition, they are often key molecules in the specific recognition mechanisms involved in both beneficial and pathogenic bacteria-host interactions. A novel EPS known as MLG (Mixed-Linkage β-Glucan) was recently discovered in rhizobia, where it participates in bacterial aggregation and biofilm formation and is required for efficient attachment to the roots of their legume host plants. MLG is the first and, so far, the only reported linear Mixed-Linkage β-glucan in bacteria, containing a perfect alternation of β (1 → 3) and β (1 → 4) bonds. A phylogenetic study of MLG biosynthetic genes suggests that far from being exclusive of rhizobia, different soil and plant-associated bacteria likely produce MLG, adding this novel polymer to the plethora of surface polysaccharides that help bacteria thrive in the changing environment and to establish successful interactions with their hosts.
In this work, a quantification method for MLG is proposed. It relays on the hydrolysis of MLG by a specific enzyme (lichenase), and the subsequent quantification of the released disaccharide (laminaribiose) by the phenol-sulfuric acid method. The protocol has been set up and optimized for its use in 96-well plates, which makes it suitable for high-throughput screening (HTS) approaches. This method stands out by its fast processing, technical simplicity, and capability to handle multiple samples and biological replicates at a time.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
More TT, Yadav JSS, Yan S et al (2014) Extracellular polymeric substances of bacteria and their potential environmental applications. J Environ Manag 144:1–25
Parsek MR, Fuqua C (2004) Biofilms 2003: emerging themes and challenges in studies of surface-associated microbial life. J Bacteriol 186:4427–4440
Costerton JW, Stewart PS, Greenberg EP (1999) Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322
Sharma S, Sharma S, Tiwari V (2022) Role of biofilm in host-pathogen interaction. In: Roy D (ed) A complete guidebook on biofilm study. Academic/Elsevier, London, p 227
Lund-Palau H, Turnbull AR, Bush A et al (2016) Pseudomonas aeruginosa infection in cystic fibrosis: pathophysiological mechanisms and therapeutic approaches. Expert Rev Respir Med 10:685–697
Ramey BE, Koutsoudis M, von Bodman SB, Fuqua C (2004) Biofilm formation in plant-microbe associations. Curr Opin Microbiol 7:602–609
Collins AJ, Smith TJ, Sondermann H, O’Toole GA (2020) From input to output: the Lap/c-di-GMP biofilm regulatory circuit. Annu Rev Microbiol 74:607–631
Ude S, Arnold DL, Moon CD et al (2006) Biofilm formation and cellulose expression among diverse environmental Pseudomonas isolates. Environ Microbiol 8:1997–2011
Aragón IM, Pérez-Mendoza D, Gallegos MT, Ramos C (2014) The c-di-GMP phosphodiesterase BifA is involved in the virulence of bacteria from the Pseudomonas syringae complex. Mol Plant Pathol 16:604–615
Perez-Mendoza D, Felipe A, Ferreiro MD et al (2019) AmrZ and FleQ co-regulate cellulose production in Pseudomonas syringae pv. tomato DC3000. Front Microbiol 10:746
Pérez-Mendoza D, Aragón IM, Prada-Ramírez HA et al (2014) Responses to elevated c-di-GMP levels in mutualistic and pathogenic plant-interacting bacteria. PLoS One 9:e91645
Pérez-Mendoza D, Romero-Jiménez L, Rodríguez-Carvajal MA et al (2022) The role of two linear β-glucans activated by c-di-GMP in Rhizobium etli CFN42. Biology 11:1364
Pérez-Mendoza D, Rodríguez-Carvajal MA, Romero-Jiménez L et al (2015) Novel mixed-linkage beta-glucan activated by c-di-GMP in Sinorhizobium meliloti. Proc Natl Acad Sci U S A 112:E757–E765
Fujishige NA, Kapadia NN, De Hoff PL, Hirsch AM (2006) Investigations of Rhizobium biofilm formation. FEMS Microbiol Ecol 56:195–206
Augimeri RV, Varley AJ, Strap JL (2015) Establishing a role for bacterial cellulose in environmental interactions: lessons learned from diverse biofilm-producing. Front Microbiol 6:1282
Navya PV, Gayathri V, Samanta D, Sampath S (2022) Bacterial cellulose: a promising biopolymer with interesting properties and applications. Int J Biol Macromol 220:435–461
Xu L, Zhang J (2016) Bacterial glucans: production, properties, and applications. Appl Microbiol Biotechnol 100:9023–9036
Caseiro C, Dias JNR, de Andrade Fontes CMG, Bule P (2022) From cancer therapy to winemaking: the molecular structure and applications of β-glucans and β-1,3-glucanases. Int J Mol Sci 23:3156
Goodridge HS, Wolf AJ, Underhill DM (2009) β-glucan recognition by the innate immune system. Immunol Rev 230:38–50
Rebaque D, Del Hierro I, Lopez G et al (2021) Cell wall-derived mixed-linked β1,3/1,4-glucans trigger immune responses and disease resistance in plants. Plant J 106:601–615
Anwar MI, Muhammad F, Awais MM, Akhtar M (2017) A review of β-glucans as a growth promoter and antibiotic alternative against enteric pathogens in poultry. World Poult Sci J 73:651–661
Vetvicka V, Vannucci L, Sima P (2014) The effects of β-glucan on pig growth and immunity. Open Biochem J 8:89–93
Clavaud C, Aimanianda V, Latge J-P (2009) Organization of fungal, oomycete and lichen (1,3)-β-glucans. In: Bacic A, Fincher GB, Stone BA (eds) Chemistry, biochemistry, and biology of 1–3 beta glucans and related polysaccharides. Academic, San Diego, p 387
Henrion M, Francey C, Lê K-A, Lamothe L (2019) Cereal β-glucans: the impact of processing and how it affects physiological responses. Nutrients 11:1729
Pérez-Mendoza D, Sanjuán J (2016) Exploiting the commons: cyclic diguanylate regulation of bacterial exopolysaccharide production. Curr Opin Microbiol 30:36–43
Pérez-Mendoza D, Bertinetti D, Lorenz R et al (2017) A novel c-di-GMP binding domain in glycosyltransferase BgsA is responsible for the synthesis of a mixed-linkage β-glucan. Sci Rep 7:8997
Morgan JL, McNamara JT, Zimmer J (2014) Mechanism of activation of bacterial cellulose synthase by cyclic di-GMP. Nat Struct Mol Biol 21:489–496
Ruffing AM, Chen RR (2012) Transcriptome profiling of a curdlan-producing Agrobacterium reveals conserved regulatory mechanisms of exopolysaccharide biosynthesis. Microbe Cell Fact 11:17
Lynd LR, Weimer PJ, van Zyl WH, Pretorius IS (2002) Microbial cellulose utilization: fundamentals and biotechnology. Microbiol Mol Biol Rev 66:506–577
Wood PJ, Fulcher RG (1984) Specific interaction of aniline blue with (1→3)-β-d-glucan. Carbohydr Polym 4:49–72
Evans NA, Hoyne PA, Stone BA (1984) Characteristics and specificity of the interaction of a fluorochrome from aniline blue (sirofluor) with polysaccharides. Carbohydr Polym 4:215–230
Nakanishi I, Kimura K, Suzuki T et al (1976) Demonstration of curdlan-type polysaccharide and some other β-1,3-glucan in microorganisms with aniline blue. J Gen Appl Microbiol 22:1–11
Reichhardt C, McCrate OA, Zhou X et al (2016) Influence of the amyloid dye Congo red on curli, cellulose, and the extracellular matrix in E. coli during growth and matrix purification. Anal Bioanal Chem 408:7709–7717
Zevenhuizen LP, Bertocchi C, van Neerven AR (1986) Congo red absorption and cellulose synthesis by Rhizobiaceae. Antonie Van Leeuwenhoek 52:381–386
DuBois M, Gilles KA, Hamilton JK et al (1956) Colorimetric method for determination of sugars and related substances. Anal Chem 28:350–356
Masuko T, Minami A, Iwasaki N et al (2005) Carbohydrate analysis by a phenol-sulfuric acid method in microplate format. Anal Biochem 339:69–72
Robertsen BK, Aman P, Darvill AG et al (1981) The structure of acidic extracellular polysaccharides secreted by Rhizobium leguminosarum and Rhizobium trifolii. Plant Physiol 67:389–400
Glazebrook J, Walker GC (1989) A novel exopolysaccharide can function in place of the calcofluor-binding exopolysaccharide in nodulation of alfalfa by Rhizobium meliloti. Cell 56:661–672
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Marchante, J.A., Ruiz-Sáez, L., Muñoz, S., Sanjuán, J., Pérez-Mendoza, D. (2024). Quantification of Mixed-Linkage β-Glucan (MLG) in Bacteria. In: Medina, C., López-Baena, F.J. (eds) Host-Pathogen Interactions. Methods in Molecular Biology, vol 2751. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-3617-6_9
Download citation
DOI: https://doi.org/10.1007/978-1-0716-3617-6_9
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-3616-9
Online ISBN: 978-1-0716-3617-6
eBook Packages: Springer Protocols