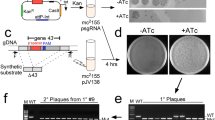
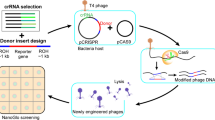
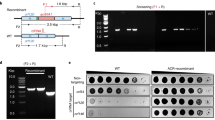
Abstract
The functional characterization of “hypothetical” phage genes is a major bottleneck in basic and applied phage research. To compound this issue, the most suitable phages for therapeutic applications—the strictly lytic variety—are largely recalcitrant to classical genetic techniques due to low recombination rates and lack of selectable markers. Here we describe methods for fast and effective phage engineering that rely upon a Type III-A CRISPR-Cas system. In these methods, the CRISPR-Cas system is used as a powerful counterselection tool to isolate rare phage recombinants.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Clokie MRJ, Millard AD, Letarov AV et al (2011) Phages in nature. Bacteriophage 1(1):31–45
Twort FW (1915) An investigation on the nature of ultra-microscopic viruses. Lancet 186(4814):1241–1243
D’Herelle F (1917) Sur Un Microbe Invisible Antagoniste Des Bacilles Dysentériques. CR Acad Sci Paris 165:373–375
Aswani VH, Shukla SK (2021) An early history of phage therapy in the United States: is it time to reconsider? Clin Med Res 19(2):82–89
Pires DP, Cleto S, Sillankorva S et al (2016) Genetically engineered phages: a review of advances over the last decade. Microbiol Mol Biol Rev 80(3):523–543
Chen Y, Batra H, Dong J et al (2019) Genetic engineering of bacteriophages against infectious diseases. Front Microbiol 10:1–12
Mahler M, Costa AR, Beljouw SPB et al (2022) Approaches for bacteriophage genome engineering. Trends Biotechnol:1–17. https://doi.org/10.1016/j.tibtech.2022.08.008
Hatoum-Aslan A (2018) Phage genetic engineering using CRISPR–Cas systems. Viruses 10(6):335
Barrangou R, Fremaux C, Deveau H et al (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712
Jansen R, van Embden JDA, Gaastra W et al (2002) Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol 43(6):1565–1575
Hille F, Richter H, Wong SP et al (2018) The biology of CRISPR-Cas: backward and forward. Cell 172(6):1239–1259
Makarova KS, Wolf YI, Iranzo J et al (2020) Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol 18:67–83
Bari SMN, Walker FC, Cater K et al (2017) Strategies for editing virulent staphylococcal phages using CRISPR-Cas10. ACS Synth Biol 6(12):2316–2325
Marraffini LA, Sontheimer EJ (2008) CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322(5909):1843–1845
Hatoum-Aslan A, Maniv I, Samai P et al (2014) Genetic characterization of Antiplasmid immunity through a type III-A CRISPR-Cas system. J Bacteriol 196(2):310–317
Maniv I, Jiang W, Bikard D et al (2016) Impact of different target sequences on type III CRISPR-Cas immunity. J Bacteriol 198(6):941–950
Hatoum-Aslan A, Samai P, Maniv I et al (2013) A ruler protein in a complex for antiviral defense determines the length of small interfering CRISPR RNAs. J Biol Chem 288(39):27888–27897
Goldberg GW, Jiang W, Bikard D et al (2014) Conditional tolerance of temperate phages via transcription-dependent CRISPR-Cas targeting. Nature 514(7524):633–637
Marraffini LA, Sontheimer EJ (2010) Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature 463(7280):568–571
Samai P, Pyenson N, Jiang W et al (2015) Co-transcriptional DNA and RNA cleavage during type III CRISPR-Cas immunity. Cell 161(5):1164–1174
Liu TY, Liu J-J, Aditham AJ et al (2019) Target preference of type III-A CRISPR-Cas complexes at the transcription bubble. Nat Commun 10:1–13
Niewoehner O, Garcia-doval C, Rostøl JT et al (2017) Type III CRISPR – Cas systems produce cyclic oligoadenylate second messengers. Nature 548(7669):543–548
Kazlauskiene M, Kostiuk G, Venclovas Č et al (2017) A cyclic oligonucleotide signaling pathway in type III CRISPR-Cas systems. Science 357:605–609
Nasef M, Muffly MC, Beckman AB et al (2019) Regulation of cyclic oligoadenylate synthesis by the Staphylococcus epidermidis Cas10-Csm complex. RNA 25:948–962
Jiang W, Samai P, Marraffini LA (2016) Degradation of phage transcripts by CRISPR-associated RNases enables type III CRISPR-Cas immunity. Cell 164:710–721
Walker FC, Chou-Zheng L, Dunkle JA et al (2017) Molecular determinants for CRISPR RNA maturation in the Cas10 – Csm complex and roles for non-Cas nucleases. Nucleic Acids Res 45(4):2112–2123
Chou-Zheng L, Hatoum-Aslan A (2019) A type III-A CRISPR-Cas system employs degradosome nucleases to ensure robust immunity. elife 8:e45393
Chou-Zheng L, Hatoum-Aslan A (2022) Critical roles for ‘housekeeping’ nucleases in type III CRISPR-Cas immunity. elife 11:e81897
Pyenson NC, Gayvert K, Varble A et al (2017) Broad targeting specificity during bacterial type III CRISPR-Cas immunity constrains viral escape. Cell Host Microbe 22:1–11
Culbertson EK, Bari SMN, Dandu S et al (2019) Draft genome sequences of staphylococcus Podophages JBug18, Pike, Pontiff, and Pabna. Microbiol Resour Announc 8(8):e00054–e00019
Cater K, Dandu VS, Bari SMN et al (2017) A novel Staphylococcus podophage encodes a unique lysin with unusual modular design. mSphere 2(2):e00040-17
Gibson DG, Young L, Chuang R-Y et al (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6(5):343–345
Abedon ST (2011) Lysis from without. Bacteriophage 1(1):46–49
Acknowledgments
A. H.-A. holds an Investigators in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund. She is also supported by an NSF/MCB CAREER award [2054755] and NIH/NIAID [R21AI156636-01].
Declaration of Interests
A. H.-A. is the inventor of a US patent relating to this work (US20180251787A1).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Hill, C.M., Hatoum-Aslan, A. (2024). Genetic Engineering of Therapeutic Phages Using Type III CRISPR-Cas Systems. In: Azeredo, J., Sillankorva, S. (eds) Bacteriophage Therapy. Methods in Molecular Biology, vol 2734. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-3523-0_18
Download citation
DOI: https://doi.org/10.1007/978-1-0716-3523-0_18
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-3522-3
Online ISBN: 978-1-0716-3523-0
eBook Packages: Springer Protocols