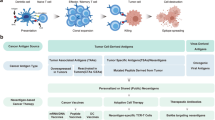
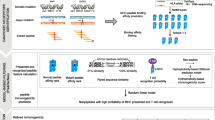
Abstract
Tumor-specific neoantigens play important roles in tumor immunotherapy. How to predict neoantigens accurately and efficiently has attracted much attention. TSNAD is the first one-stop neoantigen prediction tool from next-generation sequencing data, and TSNAdb provides both predicted and validated neoantigens based on pan-cancer immunogenomics analyses. In this chapter, we describe the usage of TSNAD and TSNAdb for the clinical application of neoantigens. The latest version of TSNAD is available at https://pgx.zju.edu.cn/tsnad, and the latest version of TSNAdb is available at https://pgx.zju.edu.cn/tsnadb.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Blass E, Ott PA (2021) Advances in the development of personalized neoantigen-based therapeutic cancer vaccines. Nat Rev Clin Oncol 18:215–229
Yamamoto TN, Kishton RJ, Restifo NP (2019) Developing neoantigen-targeted T cell–based treatments for solid tumors. Nat Med 25:1488–1499
Cui C, Wang J, Fagerberg E et al (2021) Neoantigen-driven B cell and CD4 T follicular helper cell collaboration promotes anti-tumor CD8 T cell responses. Cell 184:6101–6118.e13
Zhou Z, Lyu X, Wu J et al (2017) TSNAD: an integrated software for cancer somatic mutation and tumour-specific neoantigen detection. R Soc Open Sci 4:170050
Zhou Z, Wu J, Ren J et al (2021) TSNAD v2.0: a one-stop software solution for tumor-specific neoantigen detection. Comput Struct Biotechnol J 19:4510–4516
Zhou C, Wei Z, Zhang Z et al (2019) pTuneos: prioritizing tumor neo antigens from next-generation sequencing data. Genome Med 11:67
Hundal J, Kiwala S, McMichael J et al (2020) PVACtools: a computational toolkit to identify and visualize cancer neoantigens. Cancer Immunol Res 8:409–420
Scholtalbers J, Boegel S, Bukur T et al (2015) TCLP: an online cancer cell line catalogue integrating HLA type, predicted neo-epitopes, virus and gene expression. Genome Med 7:118
Charoentong P, Finotello F, Angelova M et al (2017) Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep 18:248–262
Tan X, Li D, Huang P et al (2020) dbPepNeo: a manually curated database for human tumor neoantigen peptides. Database 2020:baaa004
Zhou WJ, Qu Z, Song CY et al (2019) NeoPeptide: an immunoinformatic database of T-cell-defined neoantigens. Database 2019:baz128
Wu J, Zhao W, Zhou B et al (2018) TSNAdb: a database for tumor-specific neoantigens from immunogenomics data analysis. Genomics Proteomics Bioinformatics 16:276–282
Wu J, Chen W, Zhou Y et al (2022) TSNAdb v2.0: the updated version of tumor-specific neoantigen database. Genomics Proteomics Bioinformatics. https://doi.org/10.1016/j.gpb.2022.09.012
Wu J, Wang W, Zhang J et al (2019) DeepHLApan: a deep learning approach for neoantigen prediction considering both HLA-peptide binding and immunogenicity. Front Immunol 10:2559
O’Donnell TJ, Rubinsteyn A, Laserson U (2020) MHCflurry 2.0: improved pan-allele prediction of MHC class I-presented peptides by incorporating antigen processing. Cell Syst 11:42–48.e7
Jurtz V, Paul S, Andreatta M et al (2017) NetMHCpan-4.0: improved peptide–MHC class I interaction predictions integrating eluted ligand and peptide binding affinity data. J Immunol 199:3360–3368
Xia J, Bai P, Fan W et al (2021) NEPdb: a database of T-cell experimentally-validated neoantigens and pan-cancer predicted neoepitopes for cancer immunotherapy. Front Immunol 12:644637
Vigneron N, Stroobant V, Van Den Eynde BJ, Van Der Bruggen P (2013) Database of T cell-defined human tumor antigens: the 2013 update. Cancer Immun 13:15
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No. 31971371), the Key R&D Program of Zhejiang Province (Grant No. 2020C03010), and the Alibaba-Zhejiang University Joint Research Center of Future Digital Healthcare. We thank the Information Technology Center, State Key Lab of CAD&CG, and Innovation Institute for Artificial Intelligence in Medicine, Zhejiang University for the support of computing resources. We also gratefully acknowledge the TCGA Research Network for referencing the TCGA datasets, and the TCIA for referencing HLA-type data of TCGA samples.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Wu, J., Zhou, Z. (2023). TSNAD and TSNAdb: The Useful Toolkit for Clinical Application of Tumor-Specific Neoantigens. In: Reche, P.A. (eds) Computational Vaccine Design. Methods in Molecular Biology, vol 2673. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-3239-0_11
Download citation
DOI: https://doi.org/10.1007/978-1-0716-3239-0_11
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-3238-3
Online ISBN: 978-1-0716-3239-0
eBook Packages: Springer Protocols