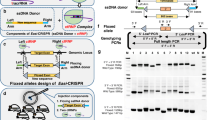
Abstract
Generation of transgenic mice by direct microinjection of foreign DNA into fertilized ova has become a routine technique in biomedical research. It remains an essential tool for studying gene expression, developmental biology, genetic disease models, and their therapies. However, the random integration of foreign DNA into the host genome that is inherent to this technology can lead to confounding effects associated with insertional mutagenesis and transgene silencing. Locations of most transgenic lines remain unknown because the methods are often burdensome (Nicholls et al., G3: Genes Genomes Genetics 9:1481–1486, 2019) or have limitations (Goodwin et al., Genome Research 29:494–505, 2019). Here, we present a method that we call Adaptive Sampling Insertion Site Sequencing (ASIS-Seq) to locate transgene integration sites using targeted sequencing on Oxford Nanopore Technologies’ (ONT) sequencers. ASIS-Seq requires only about 3 ug of genomic DNA, 3 hours of hands-on sample preparation time, and 3 days of sequencing time to locate transgenes in a host genome.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Gordon JW, Ruddle FH (1981) Integration and stable germ line transmission of genes injected into mouse Pronuclei. Science 214:1244–1246
Fielder TJ, Barrios L, Montoliu L (2010) A survey to establish performance standards for the production of transgenic mice. Transgenic Res 19:675–681
Hammer RE et al (1987) The rat elastase I regulatory element is an enhancer that directs correct cell specificity and developmental onset of expression in transgenic mice. Mol Cell Biol 8:2956–2967
Petitclerc D et al (1995) The effect of various introns and transcription terminators on the efficiency of expression vectors in various cultured cell lines and in the mammary gland of transgenic mice. Biotechnol 40:169–178
Schedl A et al (1993) A yeast artificial chromosome covering the tyrosinase gene confers copy number-dependent expression in transgenic mice. Nature 362:258–256
Ioannou PA et al (1994) A new bacteriophage P1-derived vector for the propagation of large human DNA fragments. Nat Genet 6:84–89
Yang XW, Model P, Heintz N (1997) Homologous recombination based modification in Escherichia coli and germline transmission in transgenic mice of a bacterial artificial chromosome. Nat Biotech 15:859–865
Giraldo P, Montoliu L (2001) Size matters: use of YACs BACs and PACs in transgenic animals. Transgenic Res 10:83–103
Brinster RL et al (1985) Factors affecting the efficiency of introducing foreign DNA into mice by microinjecting eggs. Proc Nail Acad Sci USA 82:4438–4442
Hirabayashi M et al (2001) A comparative study on the integration of exogenous DNA into mouse rat rabbit and pig genomes. Exp Anim 50:125–131
Palmiter RD, Brinster RL (1985) Transgenic mice. Cell 41:343–345
Garrick D et al (1998) Repeat-induced gene silencing in mammals. Nat Genet 18:56–59
Mátés L et al (2009) Molecular evolution of a novel hyperactive sleeping beauty transposase enables robust stable gene transfer in vertebrates. Nat Genetic 41:53–61
Ding S et al (2005) Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell 122:473–483
Katter K et al (2013) Transposon-mediated transgenesis transgenic rescue and tissue-specific gene expression in rodents and rabbits. FASEB J 27:930–941
Li MA et al (2011) Mobilization of giant piggyBac transposons in the mouse genome. Nucleic Acids Res 39:e148
Durkin ME et al (2001) Integration of a c-myc transgene results in disruption of the mouse Gtf2ird1 gene the homologue of the human GTF2IRD1 gene hemizygously deleted in Williams-Beuren Syndrome. Genomics 73:20–27
Mukai HY et al (2006) Transgene insertion in proximity to the c-myb gene disrupts erythroid-megakaryocytic lineage bifurcation. Mol Cell Biol 26:7953–7965
Yong CSM et al (2015) Embryonic lethality in homozygous human Her-2 transgenic mice due to disruption of the Pds5b gene. PLoS One 10:e0136817
Galvan DL et al (2009) Genome-wide mapping of piggybac transposon integrations in primary human T cells. J Immunother 32:837–844
Li MA et al (2013) The piggyBac transposon displays local and distant reintegration preferences and can cause mutations at noncanonical integration. Sites Mol Cell Biol 33:1317–1330
Saha S et al (2015) Evaluating the potential for undesired genomic effects of the piggyBac transposon system in human cells. Nucleic Acids Res 43:1770–1782
Goodwin LO et al (2019) Large-scale discovery of mouse transgenic integration sites reveals frequent structural variation and insertional mutagenesis. Genome Res 29:494–505
Nicholls PK et al (2019) Locating and characterizing a transgene integration site by nanopore sequencing. G3: Genes Genomes Genetics 9:1481–1486
Kulnane LS et al (2002) Rapid and efficient detection of transgene homozygosity by FISH of mouse fibroblasts. Mamm Genome 13:223–226
Nakanishi T et al (2002) FISH analysis of 142 EGFP transgene integration sites into the mouse genome. Genomics 80:564–574
Ohigashi I et al (2010) Identification of the transgenic integration site in immunodeficient tgσ26 human CD3σ transgenic mice. PLoS One 5:e14391
Liang Z et al (2008) Identifying and genotyping transgene integration loci. Transgenic Res 17:979–983
Haraguchi S, Nakagawara A (2009) A simple PCR method for rapid genotype analysis of the TH-MYCN transgenic mouse. PLoS One 4:e6902
Bryda EC, Bauer BA (2010) A restriction enzyme-PCR-based technique to determine transgene insertion sites. Methods Mol Biol 597:287–299
Ji Y et al (2014) Identification of the genomic insertion site of Pmel-1 TCR α and β transgenes by next-generation sequencing. PLoS One 9:e96650
Dubose AJ et al (2013) Use of microarray hybrid capture and next-generation sequencing to identify the anatomy of a transgene. Nucleic Acids Res 41:e70
Cain-Hom C et al (2017) Efficient mapping of transgene integration sites and local structural changes in Cre transgenic mice using targeted locus amplification. Nucleic Acids Res 45:e62
de Vree PJP et al (2014) Targeted sequencing by proximity ligation for comprehensive variant detection and local haplotyping. Nat Biotech 32:1019–1025
Kovaka S et al (2021) Targeted nanopore sequencing by real-time mapping of raw electrical signal with UNCALLED. Nat Biotech 39:431–441
Payne A et al (2021) Readfish enables targeted nanopore sequencing of gigabase-sized genomes. Nat Biotech 39:442–450
Pease S, Saunders TL (eds) (2011) Advanced protocols for animal transgenesis. Springer, Berlin, Heidelberg
Hofker M, van Deursen J (eds) (2011) Transgenic mouse methods and protocols. Humana Press, Totowa, NJ
Wang W et al (2008) Chromosomal transposition of PiggyBac in mouse embryonic stem cells. Proc Natl Acad Sci U S A 105:9290–9295
Johansson T et al (2010) Building a zoo of mice for genetic analyses: a comprehensive protocol for the rapid generation of BAC transgenic mice. Genesis 48:264–280
Gong S, Kus L, Heintz N (2010) Rapid bacterial artificial chromosome modification for large-scale mouse transgenesis. Nat Protoc 5:1678–1696
Beermann F et al (1993) Perinatal activation of a tyrosine aminotransferase fusion gene does not occur in albino lethal mice. Mech Dev 42:59–65
Martin S et al (2022) Nanopore adaptive sampling: a tool for enrichment of low abundance species in metagenomic samples. Genome Biol 23:11
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Yu, C. et al. (2023). ASIS-Seq: Transgene Insertion Site Mapping by Nanopore Adaptive Sampling. In: Saunders, T.L. (eds) Transgenesis. Methods in Molecular Biology, vol 2631. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-2990-1_5
Download citation
DOI: https://doi.org/10.1007/978-1-0716-2990-1_5
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-2989-5
Online ISBN: 978-1-0716-2990-1
eBook Packages: Springer Protocols