Abstract
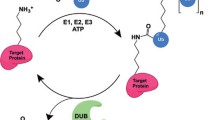
The traditional textbook describes ubiquitylation as the conjugation of ubiquitin to a target by forming a covalent bond connecting ubiquitin’s carboxy-terminal glycine residue with an acceptor amino acid like lysine or amino-terminal methionine in the substrate protein. While this adequately depicts a significant fraction of cellular ubiquitylation processes, a growing number of ubiquitin modifications do not follow this rule. Recent data demonstrate that ubiquitin can also be efficiently attached to other amino acids, such as cysteine, serine, and threonine, via ester bonding. Initially observed for a virus-encoded ubiquitin ligase, which targets a cysteine residue in a host protein to initiate its degradation, ester-linked ubiquitylation is now shown to also drive regular cellular processes. These ubiquitylation events expand the complexity and diversity of ubiquitin signaling and broaden the capability of cellular messages in the so-called ubiquitin code. Still, questions on the prevalence, relevance, and involvement in physiological and cellular functions await clearing. In this review, we aim to summarize our knowledge on ester-linked ubiquitylation and introduce experimental strategies to circumvent technical issues that complicate analysis of this uncommon posttranslational modification.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Scheffner M, Nuber U, Huibregtse JM (1995) Protein ubiquitination involving an E1–E2–E3 enzyme ubiquitin thioester cascade. Nature 373:81–83
Chau V, Tobias JW, Bachmair A et al (1989) A Multiubiquitin Chain Is Confined to Specific Lysine in a Targeted Short-Lived Protein. Science 80(243):1576–1583
Xu P, Duong DM, Seyfried NT et al (2009) Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell 137:133–145
Akutsu M, Dikic I, Bremm A (2016) Ubiquitin chain diversity at a glance. J Cell Sci 129:875–880
Komander D, Rape M (2012) The ubiquitin code. Annu Rev Biochem 81:203–229
Cadwell K, Coscoy L (2005) Ubiquitination on nonlysine residues by a viral E3 ubiquitin ligase. Science 80(309):127–130
Carvalho AF, Pinto MP, Grou CP et al (2007) Ubiquitination of mammalian Pex5p, the peroxisomal import receptor. J Biol Chem 282:31267–31272
Williams C, Van Den BM, Sprenger RR et al (2007) A conserved cysteine is essential for Pex4p-dependent ubiquitination of the peroxisomal import receptor Pex5p. J Biol Chem 282:22534–22543
Vosper JMD, McDowell GS, Hindley CJ et al (2009) Ubiquitylation on canonical and non-canonical sites targets the transciption factor neurogenin for ubiquitin-mediated proteolysis. J Biol Chem 284:15458–15468
Wang X, Herr RA, Hansen TH (2012) Ubiquitination of substrates by esterification. Traffic 13:19–24
McDowell GS, Philpott A (2013) Non-canonical ubiquitylation: Mechanisms and consequences. Int J Biochem Cell Biol 45:1833–1842
McClellan AJ, Laugesen SH, Ellgaard L (2019) Cellular functions and molecular mechanisms of non-lysine ubiquitination. Open Biol 9:190147
Kalayil S, Bhogaraju S, Bonn F et al (2018) Insights into catalysis and function of phosphoribosyl-linked serine ubiquitination. Nature 557:734–738
Tokarev AA, Munguia J, Guatelli JC (2011) Serine-threonine ubiquitination mediates downregulation of BST-2/tetherin and relief of restricted virion release by HIV-1 Vpu. J Virol 85:51–63
Gu H, Fada BJ (2020) Specificity in ubiquitination triggered by virus infection. Int J Mol Sci 21(11):4088
Breitschopf K, Bengal E, Ziv T et al (1998) A novel site for ubiquitination: the N-terminal residue, and not internal lysines of MyoD, is essential for conjugation and degradation of the protein. EMBO J 17:5964–5973
Dittmar G, Winklhofer KF (2020) Linear ubiquitin chains: cellular functions and strategies for detection and quantification. Front Chem 7:915
Peter DM, Vögeli B, Cortina NS et al (2016) A chemo-enzymatic road map to the synthesis of CoA esters. Molecules 21(4):517
Plechanovova A, Jaffray EG, Tatham MH et al (2012) Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature 489:115
Ravid T, Hochstrasser M (2007) Autoregulation of an E2 enzyme by ubiquitin-chain assembly on its catalytic residue. Nat Cell Biol 94(9):422–427
Song J, Wang J, Jozwiak AA et al (2009) Stability of thioester intermediates in ubiquitin-like modifications. Protein Sci 18:2492–2499
De Cesare V, Carbajo Lopez D, Mabbitt PD et al (2021) Deubiquitinating enzyme amino acid profiling reveals a class of ubiquitin esterases. Proc Natl Acad Sci U S A 118:e2006947118
Creasy DM, Cottrell JS (2004) Unimod: Protein modifications for mass spectrometry. Proteomics 4:1534–1536
Udeshi ND, Mertins P, Svinkina T et al (2013) Large-scale identification of ubiquitination sites by mass spectrometry. Nat Protoc 8:1950
Udeshi ND, Svinkina T, Mertins P et al (2013) Refined preparation and use of anti-diglycine remnant (k-ε-gg) antibody enables routine quantification of 10,000s of ubiquitination sites in single proteomics experiments. Mol Cell Proteomics 12:825–831
Udeshi ND, Mani DC, Satpathy S et al (2020) Rapid and deep-scale ubiquitylation profiling for biology and translational research. Nat Commun 11(1):359
Wagner SA, Beli P, Weinert BT et al (2012) Proteomic analyses reveal divergent ubiquitylation site patterns in murine tissues. Mol Cell Proteomics 11:1578–1585
Akimov V, Barrio-Hernandez I, Hansen SVF et al (2018) UbiSite approach for comprehensive mapping of lysine and N-terminal ubiquitination sites. Nat Struct Mol Biol 25:631–640
Clague MJ, Urbé S, Komander D (2019) Breaking the chains: deubiquitylating enzyme specificity begets function. Nat Rev Mol Cell Biol 20:338–352
Jarosch E, Lenk U, Sommer T (2003) Endoplasmic reticulum-associated protein degradation. Int Rev Cytol 223:39–81
Shimizu Y, Okuda-Shimizu Y, Hendershot LM (2010) Ubiquitylation of an ERAD substrate occurs on multiple types of amino acids. Mol Cell 40:917–926
Boban M, Ljungdahl PO, Foisner R (2015) Atypical ubiquitylation in yeast targets lysine-less Asi2 for proteasomal degradation. J Biol Chem 290:2489–2495
Weber A, Cohen I, Popp O et al (2016) Sequential poly-ubiquitylation by specialized conjugating enzymes expands the versatility of a quality control ubiquitin ligase. Mol Cell 63:827–839
Lips C, Ritterhoff T, Weber A et al (2020) Who with whom: functional coordination of E2 enzymes by RING E3 ligases during poly-ubiquitylation. EMBO J 39(22):e104863
Wang X, Herr RA, Rabelink M et al (2009) Ube2j2 ubiquitinates hydroxylated amino acids on ER-associated degradation substrates. J Cell Biol 187:655–668
Tan JME, Cook ECL, Van Den Berg M et al (2019) Differential use of E2 ubiquitin conjugating enzymes for regulated degradation of the rate-limiting enzymes HMGCR and SQLE in cholesterol biosynthesis. Atherosclerosis 281:137–142
Chua NK, Hart-Smith G, Brown AJ (2019) Non-canonical ubiquitination of the cholesterol-regulated degron of squalene monooxygenase. J Biol Chem 294:8134–8147
Ishikura S, Weissman AM, Bonifacino JS (2010) Serine residues in the cytosolic tail of the T-cell antigen receptor α-chain mediate ubiquitination and endoplasmic reticulum-associated degradation of the unassembled protein. J Biol Chem 285:23916–23924
Burr ML, Cano F, Svobodova S et al (2011) HRD1 and UBE2J1 target misfolded MHC class I heavy chains for endoplasmic reticulum-associated degradation. Proc Natl Acad Sci U S A 108:2034–2039
Golnik R, Lehmann A, Kloetzel PM et al (2016) Major histocompatibility complex (MHC) class i processing of the NY-ESO-1 antigen is regulated by Rpn10 and Rpn13 Proteins and immunoproteasomes following non-lysine ubiquitination. J Biol Chem 291:8805–8815
Yuan L, Hassan BA (2014) Neurogenins in brain development and disease: an overview. Arch Biochem Biophys 558:10–13
McDowell GS, Kucerova R, Philpott A (2010) Non-canonical ubiquitylation of the proneural protein Ngn2 occurs in both Xenopus embryos and mammalian cells. Biochem Biophys Res Commun 400:655–660
Roark R, Itzhaki L, Philpott A (2012) Complex regulation controls Neurogenin3 proteolysis. Biol Open 1:1264–1272
Gilkerson J, Kelley DR, Tam R et al (2015) Lysine residues are not required for proteasome- mediated proteolysis of the auxin/indole acidic acid protein IAA1. Plant Physiol 168:708–720
Peeler JC, Schedin-Weiss S, Soula M et al (2017) Isopeptide and ester bond ubiquitination both regulate degradation of the human dopamine receptor 4. J Biol Chem 292:21623–21630
Skieterska K, Rondou P, Lintermans B et al (2015) KLHL12 Promotes Non-Lysine Ubiquitination of the dopamine receptors D4.2 and D4.4, but not of the ADHD-associated D4.7 variant. PLoS One 10(12):e0145654
Kelsall IR, Zhang J, Knebel A et al (2019) The E3 ligase HOIL-1 catalyses ester bond formation between ubiquitin and components of the Myddosome in mammalian cells. Proc Natl Acad Sci U S A 116:13293–13298
Petrova T, Zhang J, Nanda SK et al (2021) HOIL-1-catalysed, ester-linked ubiquitylation restricts IL-18 signaling in cytotoxic T cells but promotes TLR signalling in macrophages. FEBS J 288:5909–5924
Carvajal AR, Grishkovskaya I, Diaz CG et al (2021) The linear ubiquitin chain assembly complex (LUBAC) generates heterotypic ubiquitin chains. Elife 10:1–28
Platta HW, El MF, Bäumer BE et al (2009) Pex2 and Pex12 function as protein-ubiquitin ligases in peroxisomal protein import. Mol Cell Biol 29:5505–5516
Okumoto K, Misono S, Miyata N et al (2011) Cysteine ubiquitination of PTS1 receptor Pex5p regulates Pex5p recycling. Traffic 12:1067–1083
Schwartzkopff B, Platta HW, Hasan S et al (2015) Cysteine-specific ubiquitination protects the peroxisomal import receptor Pex5p against proteasomal degradation. Biosci Rep 35:1–12
Debelyy MO, Platta HW, Saffian D et al (2011) Ubp15p, a ubiquitin hydrolase associated with the peroxisomal export machinery. J Biol Chem 286:28223–28234
Hensel A, Beck S, El MF et al (2011) Cysteine-dependent ubiquitination of Pex18p is linked to cargo translocation across the peroxisomal membrane. J Biol Chem 286:43495–43505
El MF, Brinkmeier R, Schrötter A et al (2013) Distinct ubiquitination cascades act on the peroxisomal targeting signal type 2 co-receptor pex18p. Traffic 14:1290–1301
Tait SWG, De VE, Maas C et al (2007) Apoptosis induction by Bid requires unconventional ubiquitination and degradation of its N-terminal fragment. J Cell Biol 179:1453–1466
Pao KC, Wood NT, Knebel A et al (2018) Activity-based E3 ligase profiling uncovers an E3 ligase with esterification activity. Nature 556:381–385
Mabbitt PD, Loreto A, Déry MA et al (2020) Structural basis for RING-Cys-Relay E3 ligase activity and its role in axon integrity. Nat Chem Biol 16:1227–1236
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Ferri-Blazquez, A., Jarosch, E., Sommer, T. (2023). Thioester and Oxyester Linkages in the Ubiquitin System. In: Rodriguez, M.S., Barrio, R. (eds) The Ubiquitin Code. Methods in Molecular Biology, vol 2602. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-2859-1_1
Download citation
DOI: https://doi.org/10.1007/978-1-0716-2859-1_1
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-2858-4
Online ISBN: 978-1-0716-2859-1
eBook Packages: Springer Protocols