Abstract
A number of protein toxins bind at the surface of mammalian cells and after endocytosis traffic to the endoplasmic reticulum, where the toxic A chains are liberated from the holotoxin. The free A chains are then dislocated, or retrotranslocated, across the ER membrane into the cytosol. Here, in contrast to ER substrates destined for proteasomal destruction, they undergo folding to a catalytic conformation and subsequently inactivate their cytosolic targets. These toxins therefore provide toxic probes for testing the molecular requirements for retrograde trafficking, the ER processes that prepare the toxic A chains for transmembrane transport, the dislocation step itself and for the post-dislocation folding that results in catalytic activity. We describe here the dislocation of ricin A chain and Shiga toxin A chain, but also consider cholera toxin which bears a superficial structural resemblance to Shiga toxin. Recent studies not only describe how these proteins breach the ER membrane, but also reveal aspects of a fundamental cell biological process, that of ER-cytosol dislocation.
Similar content being viewed by others
Keywords
- Ricin
- Shiga toxin
- Endoplasmic reticulum (ER)
- Retrotranslocation/dislocation
- ER-associated protein degradation (ERAD)
- Protein refolding
- Chaperones
- Retrograde transport
1 Introduction
The endoplasmic reticulum (ER) lumen is the site of entry, via the Sec61-associated translocon, of nascent unfolded proteins destined for the secretory pathway or for insertion into the ER membrane. Once inside the ER, these proteins fold and mature. This may require core N-glycosylation, disulphide bond formation and oligomerisation. These processes are overseen by ER quality control surveillance which may involve a combination of retrieval from post-ER compartments, ER retention, autophagy and/or ER associated protein degradation (ERAD) mechanisms (Ellgaard et al. 1999).
ERAD comprises multiple disposal processes that recognise terminally misfolded, unwanted and orphan proteins and remove them from the ER by directing them to the cytosolic proteasomes for their destruction (Brodsky and McCracken 1999). Recognition occurs via scrutiny of N-glycosylation status (Jakob et al. 1998) and by ER chaperones (Brodsky et al. 1999) which identify misfolded and unassembled proteins, and maintain their solubility before removal. Removal (or dislocation) requires membrane-integral ubiquitin ligase complexes that normally polyubiquitylate the target ERAD substrates as they are extruded through the ‘dislocon’ to enter the cytosol (Carvalho et al. 2006; Denic et al. 2006). Polyubiquitylation provides tags both for the AAA-ATPase Cdc48p complex which acts as a cytosolic extraction motor (Bays et al. 2001; Jarosch et al. 2002; Rabinovich et al. 2002; Ye et al. 2001) and for subsequent interactions with the proteasome (Elkabetz et al. 2004; Lipson et al. 2008). Typically, dislocated proteins are de-glycosylated if required, de-ubiquitylated and destroyed by proteasomal degradation (Vembar and Brodsky 2008).
A number of proteins are thought to utilise ERAD components to gain cytosolic access, but uncouple from the final destructive steps of ERAD. The best characterised of these are the catalytically active A chains of some plant and bacterial AB toxins which bind cell surface components and are endocytosed, subsequently trafficking to the ER lumen where the toxic A and cell-binding B chain(s) are separated. The A chains are then unfolded to cross the ER membrane, subsequently folding to a catalytic conformation to modify their cytosolic targets (Spooner et al. 2006). The yeast viral AB toxin K28 also dislocates from the ER and its A chain recovers catalytic activity in the cytosol (Heiligenstein et al. 2006). Similarly, the human hepatitis E virus ORF2 protein which is initially inserted into the ER lumen subsequently accumulates in the cytosol of infected cells (Surjit et al. 2007).
This phenomenon of ERAD-like dislocation and subsequent folding in the cytosol extends beyond cytotoxic and viral proteins. Both firefly and Renilla luciferases are taken up macropinocytotically by dendritic cells and are then thought to be trafficked in vesicular structures to the ER, where they unfold to utilise ERAD components, subsequently refolding in the cytosol (Giodini and Cresswell 2008). In addition, calreticulin, a protein that is normally regarded as an ER resident, has been identified in mammalian cytosolic extracts where it appears to have a role in integrin binding. This cytosolic pool of calreticulin is derived from an ER population and so has presumably been dislocated in an ERAD-like manner and then been refolded in the cytosol rather than being destroyed (Afshar et al. 2005). Thus an uncoupling from the final destructive stages of ERAD may be a fundamental cellular pathway for which, to date, only a few substrates have been identified. Seen in this light, ERAD pathways may be described as a sub-set of ER dislocation events that results in the destruction of the dislocating protein.
The protein toxins that utilise ERAD pathway components for cytosolic entry provide toxic probes to test for both dislocation requirements and also the processes that are required for recovery of catalytic activity in the cytosol. Here we consider mainly the ER-trafficking ricin and Shiga toxins, but research on these has not followed parallel paths. Recent research has provided substantial detail about dislocation of ricin, but details of its trafficking are sparse. In contrast, much is known about Shiga toxin trafficking, but less about the processes of its dislocation. We therefore also consider cholera toxin, which bears a superficial resemblance in structural terms to Shiga toxin. We discuss here how the use of dislocating toxins reveals not only how the individual toxin A chains themselves breach the ER membrane and regain activity, but also describes features of a fundamental cell transport process.
2 Toxin Structure and Function
2.1 Ricin
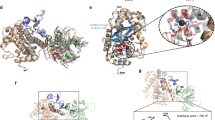
The AB toxin ricin (Fig. 1) is produced in the developing endosperm of the seeds of the castor oil plant, Ricinus communis (Harley and Beevers 1986), as a pre-proprotein that is matured by proteolytic cleavage. The mature heterodimeric holotoxin comprises a catalytically active A chain (RTA) disulphide-linked to a lectin B chain (RTB). RTA is an RNA N-glycosidase that targets the large ribosomal subunits, depurinating them by removing one specific adenyl residue from the site of interaction of elongation factors, thus halting protein synthesis (Endo and Tsurugi 1987). Since ricin maturation occurs within vacuoles derived from protein bodies of the endosperm then the host plant protein synthesis is protected from the depurination activity during germination of castor beans (Harley and Beevers 1982). Ricin B chain (RTB) binds terminal non-reducing galactose residues (exposed β1 → 4 linked galactosyls) on cell surface proteins and lipids (Olsnes et al. 1974). Whilst N-glycosylated proteins have been proposed to act as the sole potential receptors for ricin toxicity (Spilsberg et al. 2003), mammalian cells with mutant N-acetylglucosaminyl transferase 1 are unable to synthesise complex N-glycans on target proteins (Reeves et al. 2002) and yet are only protected from a ricin challenge by a factor of 20-fold (Crispin et al. 2009). This suggests that ~5% of productive receptors (those whose binding results in cytotoxicity) are non-protein, and are presumably glycolipid. To date, no definitive receptors have been identified.
Crystal structures (above) and cartoon representations (below) of the the A-B toxin ricin and the A-B5 Shiga (STx) and cholera (CTx) toxins. Crystal structures [PDB codes 2AAI (Rutenber et al. 1991), 1R4Q (Fraser et al. 2004), 1S5E (O’Neal et al. 2004), respectively] were viewed in Chimera (UCSF) and are shown from the side with the receptor-binding surfaces of the B chains (green) facing downwards. Arrowheads in the cartoons show the site of proteolytic cleavage that is required for activation of STx and CTx, separating the A chains (red) into A1 and A2 products which remain in close association, held by both non-covalent interactions and by a disulphide bond linking the two (orange). The A1 chains dislocate from the ER, leaving the A2 portion associated with the pentameric B ring. The A and B chains of ricin are held together by hydrophobic interactions and a disulphide bond
2.2 Shiga Toxin and Cholera Toxin
Shiga toxin (STx) is synthesised by the bacterium Shigella, and the closely related Shiga-like toxins by enterohemorrhagic strains of E. coli. All comprise an A1–A2 chain which is tightly associated with a doughnut-shaped ring of 5 B-chain subunits (Fig. 1). Cleavage by furin in the early stages of target mammalian cell intracellular transport results in an activated A1 chain disulphide linked to the A2 chain (Garred et al. 1995). It is the STxA1 chain that subsequently dislocates (LaPointe et al. 2005). Like RTA, STxA1 also depurinates the large ribosomal subunit, in a catalytically identical process (Endo et al. 1988). Each STxB chain subunit can bind 3 molecules of its receptor, the glycolipid globotriaosylceramide or Gb3 (Jacewicz et al. 1989). Upon cell-surface Gb3 binding extensive cross-linking of receptor forces a reorganization of the lipid membrane, a cargo-induced promotion of membrane curvature and invagination (Römer et al. 2007; Windschiegl et al. 2009).
Superficially similar in structure to STx is cholera toxin (CTx), produced by the bacterium Vibrio cholera (Fig. 1). It also has an activated A1 chain, disulphide linked to an A2 chain that remains tightly associated with a pentameric ring of B subunits. Activation, though, is by the metalloproteinase ADAM-17, rather than by furin (Valeva et al. 2004), and the A1 chain stimulates adenyl cyclase, which increases cyclic AMP concentrations in the cytosol and results in massive secretory diarrhoea (Sharp and Hynie 1971). The CTxB subunits can each bind three molecules of its lipid receptor GM1 with a similar geometry to STxB/Gb3 binding (Heyningen 1974), an energetically stable clustering that allows STxB and polyoma family viruses to induce membrane invaginations (Ewers et al. 2010).
3 Toxin Intracellular Trafficking to the Endoplasmic Reticulum
The three toxins considered here all bind their respective receptors at the plasma membrane and are internalised endocytotically, trafficking via early endosomes, the trans-Golgi network and the Golgi stack to the endoplasmic reticulum (ER) lumen (Fig. 2). By far the best characterised toxin transport details have come from study of B chains of STx, with trafficking studies of ricin and cholera toxin contributing little overall. A number of recent reviews describe and compare these trafficking events (Bonifacino and Rojas 2006; Johannes and Decaudin 2005; Johannes and Popoff 2008; Sandvig and van Deurs 2005; Spooner et al. 2006; Spooner and Watson 2010), so we will not consider this in detail, apart from describing a few recent observations that concern endosomal and Golgi passage.
Trafficking schemes. Ricin, CTx and STx bind their respective receptors at the plasma membrane and after internalization by endocytosis, traffic via early endosomes, the TGN and the Golgi stack to the ER, where the toxic polypeptides (A or A1 chains) are processed by ER chaperones (gray-blue) resulting in reductive separation from the holotoxin and maintenance of solubility prior to dislocation and recovery of activity in the cytosol. Despite the common stages in trafficking, and a common docking mechanism at the TGN for ricin and STx, the routes taken by these toxins are otherwise idiosyncratic
Although early observations concluded that STx and CTx traffic indistinguishably to the Golgi complex (Nichols et al. 2001), it has become clear that these pathways can be separated at the endosomal level by use of the small molecule Exo2, an inhibitor of a sub-set of Arf-GEF functions (Boal et al. 2010) which strongly blocks egress of STx from the early endosomes (Spooner et al. 2008b) but which has little or no effect on CTx trafficking (Feng et al. 2004). Furthermore, the clathrin associated Hcs70 co-chaperone RME-8 regulates endosomal trafficking of STx (Popoff et al. 2009) whilst having no effect on the transport of cholera toxin (Girard et al. 2005).
The role of the Golgi stack is not clear, since all three toxins negotiate this without using the COPI-dependent route that characterises retrograde Golgi transport. Golgi functions are essential, at least for STx, since subcellular surgery to remove the Golgi stack halts the toxin retrograde transport process (McKenzie et al. 2009). Nevertheless, a discrete Golgi stack is not necessary, since a derivative of Exo2 that fuses Golgi and ER membranes and disrupts the remaining Golgi into small punctae has no measurable effect on STx toxicity (Guetzoyan et al. 2010).
It appears that whilst all three toxins traffic from early endosomes to the trans-Golgi network and through the Golgi stack, the routes taken are idiosyncratic. Nevertheless, the terminal destination of holotoxin is the ER lumen (Rapak et al. 1997; Sandvig et al. 1994; Wales et al. 1992).
4 ER Pre-dislocation Events
ERAD substrates are recognised as terminally unfolded and then dispatched to the cytosol for destruction. However, ER-trafficking toxins arrive in the ER as fully-folded stable molecules and are thus very poor substrates for dislocation processes. The key to their toxicity, then, lies in destabilisation of the toxins upon ER luminal arrival by reductive separation of the catalytic A or A1 chains from their ER-targeting B chains, followed by unfolding and recognition as substrates for dislocation.
4.1 Reductive Separation of the Holotoxin Subunits
In the absence of heat and denaturant, high concentrations of the small molecule reducing agent DTT are required to separate the RTA and RTB chains of ricin in vitro (Emmanuel et al. 1988; Simpson et al. 1995c): thus the disulphide bridge connecting these chains is occluded, and separation would normally require a remodelling of the holotoxin structure to open the interface between RTA and RTB and allow reductive cleavage. Protein disulphide isomerase (PDI) possesses the qualities required, since it acts both as a protein-binding chaperone and as a disulphide exchanger (Ferrari and Söling 1999; Klappa et al. 1997). Upon separation of the A and B chains, the nearest RTA binding partner with an available oxidisable cysteine would be its erstwhile RTB partner (Fig. 2). Thus, overexpression of RTB at the site of reduction should protect cells from ricin by acting as a dead-end receptor for newly-released RTA. Expression of RTB in the mammalian ER has precisely this effect (Spooner et al. 2004). The ER-expressed RTB is retained for some time in the ER by a thiol anchor constituted by a mixed disulphide between RTB and PDI. Breaking this bond in vivo by treating the cells with DTT reverses the protective effect of RTB expression. Furthermore, in vitro, PDI can reduce ricin in the presence of thioredoxin, thioredoxin reductase and NADPH and in vivo, auranofin, an irreversible inactivator of thioredoxin reductase protects cells against challenge with ricin but not with pre-reduced ricin (Bellisola et al. 2004). Taken together, these experiments point to a physiological role for PDI in ricin cytotoxicity.
Since the A1 and A2 chains of SLTx are disulphide linked, then a reductive event must be required prior to dislocation of SLTxA1, but to date, this has not been characterised. However, results that rationalised what had previously been a quandary—how a disulphide can be cleaved in the relatively oxidising environment of the ER lumen (Majoul et al. 1996)—showed that for CTx, the bridge between the A1 and A2 chains is sensitive to the ratio of oxidised and reduced glutathione in the ER and that ER-lumenal PDI is required for the acceleration of the reduction of CTxA1 and A2 (Majoul et al. 1997; Orlandi 1997).
4.2 Unfolding of the Toxin A Chains
Comparison of crystal structures of ricin and RTA reveals that RTA takes up different stable conformations depending upon whether it is part of holotoxin or not. Thus reductive separation of RTA and RTB results in a general re-modelling of RTA, a specific displacement of its C-terminal tail (Fig. 3, cyan) and the exposure of a relatively hydrophobic patch that had previously been occluded by RTB (Fig. 3, blue). A number of mutations in this region have no major effect on catalytic activity of RTA in vitro, nor on in vitro re-association of recombinant RTA with wild-type RTB, and yet the reassociated holotoxins display reduced toxicity (Simpson et al. 1995b). Similarly, adding charged residues at the C-terminus of RTA hinders toxicity without obvious effects on catalytic activity or subunit re-association/dissociation (Simpson et al. 1995c).
Reductive separation of RTA and RTB results in a re-modelling of RTA structure. a, c structures of ricin (Rutenber et al. 1991) and b, d, e RTA, PDB code 2VC4 (Allen et al. 2007), displayed as ribbons (a, b) and space-fill (c–e). Upon separation, a hydrophobic patch (blue) of RTA (red), formerly occluded by RTB (green) is revealed, best seen in a view from below RTA (e) and the extreme C-terminus of RTA (cyan) is displaced
Taken together, these results implicate the hydrophobic stretch as important for membrane translocation, suggesting that this region interacts with the ER membrane prior to RTA dislocation. Supporting this interpretation is the partitioning of RTA, but not holoricin, in the detergent phase after Triton-X114 extraction, and the spontaneous structural changes that occur in RTA in the presence of negatively charged phospholipids (Day et al. 2002; Mayerhofer et al. 2009). Furthermore, when RTA is tagged with fluorophores in different positions around the molecule, some fluorophore tags are quenched in the presence of microsomal membranes pre-soaked with a lipopholic quencher, whilst fluorophores in other positions are not. Thus the interaction with lipid membranes is not random—some parts of the RTA molecule are excluded from the lipid bilayer, whilst at least two amino acids in the hydrophobic C-terminal patch of RTA (Cys259 and I249) enter the non-polar core of a lipid membrane (Mayerhofer et al. 2009). Insertion into the membrane is temperature-dependent: at low temperatures, RTA binds membranes, and as the temperature increases, structural changes associated with membrane entry become more apparent (Mayerhofer et al. 2009). Isolated purified RTA is temperature-sensitive, forming a molten globule at 45°C (Argent et al. 2000), but even at 37°C, it is relatively unstable, and prone to aggregation (Spooner et al. 2008a).
The driving force for RTA unfolding thus appears to be thermal instability of RTA released from holotoxin coupled with an ordered insertion into the membrane that results in a remodelling of RTA structure. This membrane-embedded form is thought to mimic a misfolded protein that is then dislocated from the ER in an ERAD-like manner.
The A1 chain of STx also possesses a relatively hydrophobic string of amino-acids, which is required for the dislocation of the A1 chain expressed in the yeast ER (LaPointe et al. 2005). Furthermore, an artificial peptide based on this sequence interacts with lipid membranes (Menikh et al. 1997; Saleh et al. 1996). Whilst the parallels with RTA structure might seem obvious, we note that this region is already substantially exposed in the holotoxin (Fig. 4, blue), and it remains to be determined whether membrane interactions induce conformational changes in SLTxA1 that are then recognized by the dislocation machinery.
An examination of the CTxA1 chain shows no obvious candidates for a hydrophobic region exposed after subunit separation that may act to destabilise A1 structure by interacting with lipid membranes. Instead, an overt role for PDI in the unfolding of the A1 chain has been proposed, since in the presence of reduced PDI, the CTxA1 chain becomes markedly trypsin sensitive (Tsai et al. 2001). Another member of the PDI family, ERp72, plays an ER retention role for CTxA, stabilising its structure in a trypsin-resistant form (Forster et al. 2006). An alternative model suggests that unfolding of CTxA occurs because it is, like RTA, thermally unstable after release from holotoxin (Pande et al. 2007), with a protease-sensitive structure at 37°C that is not apparent in the holotoxin. Thermal instability in the dissociated CTxA1 chain could thus allow it to mimic a misfolded protein for ERAD-like export to the cytosol. Consistent with this, in vivo stabilisation of the structure of CTxA1 with glycerol reduces cholera toxicity by inhibiting dislocation (Massey et al. 2009). Furthermore, dissection of the process of unfolding reveals subdomains of A1 that are temperature-sensitive and those that serve to stabilise A1 structure, studies that may lead to a full mechanistic description of the unfolding process (Banerjee et al. 2010).
4.3 Maintenance of Solubility and Recognition as Substrates for ER-cytosol Dislocation
Hydrophobic regions on otherwise soluble proteins provide motifs for recognition by ER chaperones such as the Hsc70 family chaperone BiP, which maintain solubility of these substrates (Brodsky et al. 1999). The initial remodelling of RTA after subunit reduction may therefore explain why, when expressed in the yeast ER, maximal toxicity of RTA requires the co-chaperones of Kar2p, the yeast equivalent of BiP (Li et al. 2010) and rationalise a proposed role for the Hsp90 family ER chaperone GRP94 in ricin toxicity to mammalian cells (Spooner et al. 2008a; Taylor et al. 2010).
The process of recognition of an ERAD substrate is currently an active area of research, using only a few known model substrates. For example, specific N-glycans appear to act as folding sensors that can recognise altered protein stability (Spear and Ng 2005; Xie et al. 2009). These signals are read by the EDEM (mammalian) and Yos9p (yeast) families of lectin chaperones (Kanehara et al. 2007). A role for EDEM has been proposed in transporting free RTA to the ‘dislocon’ for ER removal (Slominska-Wojewodzka et al. 2006), but since ricin constituted with a recombinant (unglycosylated) RTA is as toxic as plant-derived ricin (with a glycosylated RTA), then a direct role through N-glycan recognition would seem unlikely. Furthermore there is no obvious role for Htm1p, the yeast equivalent of EDEM, when RTA is expressed in the yeast ER lumen (Li et al. 2010), nor for any other molecules typically regarded as part of glycoprotein surveillance. However, EDEM is part of a three-subunit troika with BiP and Erdj5 (an Hsp40 co-chaperone of BiP with PDI activity) that has roles in ERAD (Ushioda et al. 2008), so manipulating EDEM levels may disturb this whole complex.
When expressed in the yeast ER, RTA dislocation is preceded by engagement with a specific COPII-interacting ER transmembrane p24 protein, leading to possible Golgi trafficking and subsequent return to the ER (Li et al. 2010). This may be a universal requirement for ricin toxicity, since partial screening of an RNAi library in Drosophila melanogaster S2 cells for sensitivity changes to ricin challenge also identifies the fly equivalent of Erp2p and its interacting proteins as important in ricin toxicity (Pawar et al. 2011). Furthermore, in mammalian cells, ricin cytotoxicity is perturbed by interfering with Rab1 and Sar1, which control ER to Golgi trafficking (Simpson et al. 1995a), again suggesting a requirement for RTA to enter the Golgi from the ER. Thus recognition of RTA as a misfolded substrate for dislocation may not occur in the ER lumen, but perhaps in the Golgi or at least in the context of proteins that associate with COPII coated buds. The final recognition of misfolded RTA as a dislocatable substrate appears to be by membrane-integral components of the HRD ubiquitin ligase complex that probably forms the dislocon.
For STx, the hydrophobic stretch in the A1 subunit is already substantially exposed in the holotoxin (Fig. 3, blue), so the strong interactions of STxA with a pre-assembled ER luminal protein complex containing the chaperones HEDJ, BiP and GRP94 associated with the Sec61 translocon core unit (Falguieres and Johannes 2006; Yu and Haslam 2005) may also be initiated with the holotoxin. BiP is also thought to maintain the solubility of CTxA1 (Winkeler et al. 2003).
5 Dislocation Across the ER Membrane
Currently, the RTA dislocation process is largely uncharacterised in mammalian cells: little is known beyond the implication of Sec61 as part of a dislocation channel (Simpson et al. 1999; Slominska-Wojewodzka et al. 2006; Wesche et al. 1999) and the lack of obvious function of derlins (Slominska-Wojewodzka et al. 2006).
However, when expressed exogenously in the yeast ER lumen RTA dislocates and folds to an active conformation in the cytosol, reducing yeast protein synthesis activity and causing a severe growth defect (Li et al. 2010). This has allowed us to examine the requirements for RTA dislocation (Fig. 5).
Dislocation of RTA expressed exogenously in the yeast ER lumen. 1 Prior to dislocation there is a compulsory interaction of RTA with a COPII associated p24 protein (Erp2p) that may point to a requirement for ER-Golgi transport for recognition or retrieval of misfolded RTA. 2 Dislocation requires the core components of the Hrd1p–Hrd3p dislocaon, but not the Hrd1p E3 ubiquitin ligase activity encoded by its RING-H2 domain (H2). The requirements for der1p, Usa1p, Cue1p and Ubc7p are intermediate and for Ubx2p, the requirement is minor. 3 Extraction of RTA from the dislocon requires the Rpt4p subunit of the proteasome cap. 4 In contrast, the mutant RTAΔ that is unable to fold to a catalytic conformation is a bonafide ERAD substrate that is ubiquitylated by Hrd1p and extracted by the Cdc48 complex, deglycosylated by Png1p and requires the proteasomal receptor Rad23p and the proteasomal cap unit Rpt2 which provides access to the proteolytic activities of the proteasome core
RTA utilises the integral membrane protein HRD E3 ubiquitin ligase complex for dislocation. This comprises the multi-spanning Hrd1p ubiquitin ligase which may constitute the central core of a dislocon (Gauss et al. 2006), the Hrd3p protein that is required for maintenance of Hrd1p (Gardner et al. 2000) and Usa1p, which promotes optimal Hrd1p activity (Carroll and Hampton 2010). This complex recognises misfolded membrane spanning domains, but has a modular design (Kanehara et al. 2010), allowing add-on functions such as those provided by Der1p and Usa1p, which adapt the complex for recognition of misfolded luminal domains (Carvalho et al. 2006), and provision for engagement via Hrd3p of proteins such as Yos9p involved in N-glycan surveillance (Denic et al. 2006). It is anchored at the dislocation site by the membrane-integral Cue1p which recruits (Biederer et al. 1997) and activates (Bazirgan and Hampton 2008) the ubiquitin-conjugating E2 Ubc7p. RTA has relatively minor requirements for Usa1p, Cue1p and Ubx7p and shows no requirements for Yos9p functions. Furthermore, RTA dislocation is independent of the E3 activity of Hrd1p (Li et al. 2010). ER dislocation of RTA in plant cells is also independent of canonical ubiquitylation (Marshall et al. 2008). Consistent with the lack of dislocation-associated polyubiquitylation, RTA is not extracted from the yeast ER by Cdc48p or its ubiquitin-handling co-factors (Li et al. 2010). Instead the extraction motor appears to be the Rpt4p subunit of the proteasomal cap, which had previously been shown to play a role in extraction of other substrates in conjunction with Cdc48p (Lipson et al. 2008). Finally, dislocated RTA is not degraded by the proteasomal core (Li et al. 2010).
Thus RTA appears to be dislocated promiscuously through non-anchored, non-optimised dislocons in a manner that either displaces the RING-H2 ubiquitin ligase domain of Hrd1p or avoids it altogether, and newly-dislocated RTA avoids proteasomal destruction. In marked contrast (Fig. 5), a mutated form of RTA that is unable to fold to an active conformation acts as a bonafide ubiquitylated and N-glycosylated ERAD substrate that is extracted in a Cdc48p-dependent manner, de-glycosylated by Png1p, passed to the proteasome by Rad21p and is then degraded by the proteasomal core (Li et al. 2010).
For STxA1 chain, dislocation has been proposed to follow the binding of chaperones which pass the protein to a complex containing the Sec61 protein (LaPointe et al. 2005). To date, molecular details remain unexplored. For CTx, mammalian cell challenge with the toxin results in up-regulation of a number of proteins involved in dislocation, including Derlin-1 (the human equivalent of yeast Der1p), gp78 and Hrd1 (Dixit et al. 2008), interpreted as up-regulation by cholera toxin of proteins required for the dislocation of its own A1 chain. CTx binds both gp78 and Hrd1 and since these are E3 ubiquitin ligases, a role for ubiquitin in dislocation has been proposed (Bernardi et al. 2010). However, like RTA, wild-type CTxA dislocates in a manner independent of canonical ubiquitylation (Rodighiero et al. 2002) and N-terminal extension of CTxA results in its conversion to an ERAD substrate by displacing the two lysyl residues normally near the N terminus, making them substrates for dislocation-associated polyubiquitylation (Wernick et al. 2010). An initial view that Derlin-1 is required for CTxA1 dislocation (Bernardi et al. 2008) has been reversed after taking into account the effect of manipulating Derlin-1 levels in the absence of CTx challenge (Saslowsky et al. 2010). Whether ER extraction requires the mammalian equivalent of Cdc48p, p97 (Abujarour et al. 2005) or not (Kothe et al. 2005) is unclear, although down-regulation of Ufd1 and Npl4, co-factors of p97, appears to sensitise cells to cholera toxin challenge (McConnell et al. 2007). A role for the cytosolic chaperone Hsp90 has also been proposed for the dislocation of CTxA, since in the presence of an Hsp90 inhibitor, there is little if any CTxA chain found in the cytosol (Taylor et al. 2010).
Dislocation of A chains might be expected to leave an excess of B chains accumulating in the ER. However, when RTB is expressed exogenously in the mammalian ER, it is trapped for a while by a thiol anchor, and then disappears from the ER by two mechanisms: approximately half is secreted, whilst the remainder becomes an ERAD substrate that can be stabilised by proteasomal inhibition (Spooner et al. 2004). The fates of STxB/A2 and CTxB/A2 are currently unmapped.
6 Cytosolic Post-dislocation Events that Restore Catalytic Activity
The protein toxins are thought to enter the cytosol in a substantially disordered conformation, so recovery of catalytic activity is expected to require extensive folding in the cytosol.
In vitro, RTA carefully unfolded to a molten globule structure can recover catalytic activity in the presence of substrate ribosomes (Argent et al. 2000). In vivo, though, in mammalian cells postdislocation scrutiny by the cytosolic chaperone Hsc70 (Fig. 6) is required for RTA to gain a catalytic conformation (Spooner et al. 2008a), followed by specific depurination of the ribosomal targets (Endo et al. 1987) and subsequent cell death. Since Hsc70 in vitro can prevent aggregation of heat denatured RTA, one in vivo role may be to aid solubility of dislocated RTA, so that it can then undergo substrate-mediated refolding. Alternatively, the role of Hsc70 may be to stabilise RTA in the cytosol by masking the hydrophobic patch that interacts with membranes. The ability of Hsc70 to aid RTA activation depends on the prevailing concentrations of Hsc70 co-chaperones, with some (HIP and BAG-2) promoting folding of RTA, and others such as the proteasome-engaging BAG-1 promoting inactivation. In addition, there is a sequential chaperone triage in the cytosol, where a proportion of RTA is passed, via the Hsc70–Hsp90 organising protein HOP, to the Hsp90 chaperone. From here, the net fate of RTA is inactivation. Although RTA is not ubiquitylated during dislocation, a low level of cytosolic ubiquitylation of RTA does occur via an unknown E3 ligase (Li et al. 2010). In vitro, RTA can be ubiquitylated very inefficiently in the presence of Hsc70 and the cytosolic CHIP E3 ubiquitin ligase, and this can be improved by mimicking the sequential triage by adding the co-chaperone HOP and the heat shock chaperone Hsp90, suggesting that Hsp90 interactions inactivate RTA by promoting cytosolic ubiquitylation (Spooner et al. 2008a). Thus a network of chaperones determines RTA fate in the cytosol by regulating the competing processes of folding and ubiquitin-tagging. This may provide a rationale for why inhibition of proteasomal degradation sensitises cells slightly to intoxication by a toxin that is not ubiquitylated during dislocation (Deeks et al. 2002; Wesche et al. 1999).
Post-dislocation scrutiny by a network of chaperones and co-chaperones determines the cytosolic fate of RTA. Non-native dislocated RTA is loaded onto Hsc70 by an Hsp40 family member (J protein). From this chaperone-bound state, routes lead to activation (folding) and inactivation (presumably by proteasome engagement). The Hsc70-interacting protein (HIP) stabilizes the Hsc70:RTA interaction, and release of RTA from this complex by BAG family guanine nucleotide exchange factors can take place in the vicinity of the proteasome (via the interlaced ubiquitin-like domain of BAG-1) or away from the proteasome (via BAG-2). Transfer of RTA from Hsc70 to Hsp90 via the Hsc70–Hsp90 operating protein HOP leads to CHIP-mediated ubiquitylation (Ub) of RTA and subsequent inactivation. In addition, there is likely to be an indirect route by direct CHIP-mediated ubiquitylation of Hsc70 that may result in proteasomal sorting of RTA (not shown)
For STx, inhibition of proteasomal degradation has been reported to give a slight increase in cytotoxicity of the toxin, of the same order of magnitude as that of sensitisation to ricin (Tam and Lingwood 2007), suggesting that there is a role for ubiquitin tagging in the fate of cytosolic STx. Thermally-unfolded CTxA can be degraded in a ubiquitin-independent fashion by the 20S core proteasome (Pande et al. 2007), so CTxA1 survival in the cytosol was thought to require rapid spontaneous folding (Rodighiero et al. 2002) allowing proteasomal avoidance. Structural studies, though, suggest that a likely route of toxin activation is via substrate-mediated folding in the cytosol with ARF6 stimulating the refolding of the C-terminus of CTx (Ampapathi et al. 2008). In addition, the requirement for Hsp90 for CTx toxicity (Taylor et al. 2010) may also reflect cytosolic post-dislocation events where Hsp90 is required for maintenance of CTxA1 in the cytosol, protecting the toxin from proteasomal degradation.
7 Concluding Remarks
From early observations that the toxic polypeptides of ER-trafficking toxins have low lysine content, and so might avoid canonical ubiquitylation and subsequent proteasomal degradation (Hazes and Read 1997), through trafficking studies that demonstrated ER arrival of holotoxin, to molecular dissection of pre-dislocation, dislocation and post-dislocation events, the notion that these toxic subunits dislocate in an ERAD-like manner has been validated, particularly for RTA.
RTA is not ubiquitylated by Hrd1p during dislocation (Li et al. 2010) but augmenting its lysyl content results in conversion to an ERAD substrate and subsequent proteasomal destruction (Deeks et al. 2002). Similarly CTxA1 dislocates in a manner independent of canonical ubiquitylation (Rodighiero et al. 2002), although appending a N-terminal extension permits two lysyl residues to become ubiquitylated, converting CTxA1 into a bonafide ERAD substrate (Wernick et al. 2010). Thus, dislocation and refolding of the wild-type toxic polypeptides must proceed in a way that protects canonical sites for ubiquitylation from the Hrd1 ubiquitin ligase activity. These toxin subunits may displace the RING-H2 domain of Hrd1. Alternatively, RTA and CTx1 may normally dislocate with a partially structured conformation that hinders access of the RING-H2 domain to these sites. Since these proteins gain catalytic activity in the cytosol, the fate of a Hrd1-dislocated protein is not necessarily destruction, and so ERAD is simply a sub-set (albeit the most studied) of ER–cytosol dislocation events. Furthermore, dislocation of RTA does not require a fully-functional or anchored dislocon, supporting the notion that there is a core dislocon that is adaptable by addition of bolt-on functions (Kanehara et al. 2010).
The key to uncoupling from the final destructive stage of ERAD, for at least the two dislocating proteins RTA and CTxA1, is avoidance of polyubiquitylation via the membrane-integral E3 ligase Hrd1. In turn this permits bypass of Cdc48/p97 interactions for cytosolic extraction and subsequent proteasomal presentation, at least for RTA. Instead RTA utilises the proteasomal cap subunit Rpt4p (Li et al. 2010), which can also provide a driving force for substrate extraction from the ER (Lipson et al. 2008). Similarly, the K28 viral killer toxin dislocates without being ubiquitylated and without assistance from Cdc48p and its Npl4p and Ufd1p co-factors, and is not degraded by the proteasomal core (Heiligenstein et al. 2006). These studies raise the question of whether or not such avoidance of Cdc48p/p97 interactions is universal for those other proteins, such as luciferases and calreticulin, which can dislocate from the ER to gain function in the cytosol.
Abbreviations
- CTx:
-
Cholera toxin
- CTxA1:
-
CTx A1 toxic chain
- CTxB:
-
CTx B chain
- ER:
-
Endoplasmic reticulum
- ERAD:
-
ER associated protein degradation
- Gb3:
-
Glycolipid globotriaosylceramide, the STx receptor.
- PDI:
-
Protein disulphide isomerise
- RTA:
-
Ricin A chain
- RTB:
-
Ricin B chain
- STx:
-
Shiga toxin
- STxA1:
-
STx A1 toxic chain
- STxB:
-
STx B chain
References
Abujarour RJ, Dalal S, Hanson PI, Draper RK (2005) p97 is in a complex with cholera toxin and influences the transport of cholera toxin and related toxins to the cytoplasm. J Biol Chem 280:15865–15871
Afshar N, Black BE, Paschal BM (2005) Retrotranslocation of the chaperone calreticulin from the endoplasmic reticulum lumen to the cytosol. Mol Cell Biol 25:8844–8853
Allen SC, Moore KA, Marsden CJ, Fülop V, Moffat KG, Lord JM, Ladds G, Roberts LM (2007) The isolation and characterization of temperature-dependent ricin A chain molecules in Saccharomyces cerevisiae. Febs J 274:5586–5599
Ampapathi RS, Creath AL, Lou DI, Craft JW Jr, Blanke SR, Legge GB (2008) Order–disorder–order transitions mediate the activation of cholera toxin. J Mol Biol 377:748–760
Argent RH, Parrott AM, Day PJ, Roberts LM, Stockley PG, Lord JM, Radford SE (2000) Ribosome-mediated folding of partially unfolded ricin A chain. J Biol Chem 275:9263–9269
Banerjee T, Pande A, Jobling MG, Taylor M, Massey S, Holmes RK, Tatulian SA, Teter K (2010) Contribution of subdomain structure to the thermal stability of the cholera toxin A1 subunit. Biochemistry 49:8839–8846
Bays NW, Wilhovsky SK, Goradia A, Hodgkiss-Harlow K, Hampton RY (2001) HRD4/NPL4 is required for the proteasomal processing of ubiquitinated ER proteins. Mol Biol Cell 12:4114–4128
Bazirgan OA, Hampton RY (2008) Cue1p is an activator of Ubc7p E2 activity in vitro and in vivo. J Biol Chem 283:12797–12810
Bellisola G, Fracasso G, Ippoliti R, Menestrina G, Rosen A, Solda S, Udali S, Tomazzolli R, Tridente G, Colombatti M (2004) Reductive activation of ricin and ricin A chain immunotoxins by protein disulfide isomerase and thioredoxin reductase. Biochem Pharmacol 67:1721–1731
Bernardi KM, Forster ML, Lencer WI, Tsai B (2008) Derlin-1 facilitates the retro-translocation of cholera toxin. Mol Biol Cell 19:877–884
Bernardi KM, Williams JM, Kikkert M, van Voorden S, Wiertz EJ, Ye Y, Tsai B (2010) The E3 ubiquitin ligases Hrd1 and gp78 bind to and promote cholera toxin retro-translocation. Mol Biol Cell 21:140–151
Biederer T, Volkwein C, Sommer T (1997) Role of Cue1p in ubiquitination and degradation at the ER surface. Science 278:1806–1809
Boal F, Guetzoyan L, Sessions RB, Zeghouf M, Spooner RA, Lord JM, Cherfils J, Clarkson GJ, Roberts LM, Stephens DJ (2010) LG186: an inhibitor of GBF1 function that causes Golgi disassembly in human and canine cells. Traffic 11:1537–1551
Bonifacino JS, Rojas R (2006) Retrograde transport from endosomes to the trans-Golgi network. Nat Rev Mol Cell Biol 7:568–579
Brodsky JL, McCracken AA (1999) ER protein quality control and proteasome-mediated protein degradation. Semin Cell Dev Biol 10:507–513
Brodsky JL, Werner ED, Dubas ME, Goeckeler JL, Kruse KB, McCracken AA (1999) The requirement for molecular chaperones during endoplasmic reticulum-associated protein degradation demonstrates that protein export and import are mechanistically distinct. J Biol Chem 274:3453–3460
Carroll SM, Hampton RY (2010) Usa1p is required for optimal function and regulation of the Hrd1p endoplasmic reticulum-associated degradation ubiquitin ligase. J Biol Chem 285:5146–5156
Carvalho P, Goder V, Rapoport TA (2006) Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell 126:361–373
Crispin M, Chang VT, Harvey DJ, Dwek RA, Evans EJ, Stuart DI, Jones EY, Lord JM, Spooner RA, Davis SJ (2009) A human embryonic kidney 293T cell line mutated at the Golgi alpha-mannosidase II locus. J Biol Chem 284:21684–21695
Day PJ, Pinheiro TJ, Roberts LM, Lord JM (2002) Binding of ricin A chain to negatively charged phospholipid vesicles leads to protein structural changes and destabilizes the lipid bilayer. Biochemistry 41:2836–2843
Deeks ED, Cook JP, Day PJ, Smith DC, Roberts LM, Lord JM (2002) The low lysine content of ricin A chain reduces the risk of proteolytic degradation after translocation from the endoplasmic reticulum to the cytosol. Biochemistry 41:3405–3413
Denic V, Quan EM, Weissman JS (2006) A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell 126:349–359
Dixit G, Mikoryak C, Hayslett T, Bhat A, Draper RK (2008) Cholera toxin up-regulates endoplasmic reticulum proteins that correlate with sensitivity to the toxin. Exp Biol Med (Maywood) 233:163–175
Elkabetz Y, Shapira I, Rabinovich E, Bar-Nun S (2004) Distinct steps in dislocation of luminal endoplasmic reticulum-associated degradation substrates: roles of endoplamic reticulum-bound p97/Cdc48p and proteasome. J Biol Chem 279:3980–3989
Ellgaard L, Molinari M, Helenius A (1999) Setting the standards: quality control in the secretory pathway. Science 286:1882–1888
Emmanuel F, Turpin E, Alfsen A, Frenoy JP (1988) Separation of ricin A and B chains after dithiothreitol reduction. Anal Biochem 173:134–141
Endo Y, Mitsui K, Motizuki M, Tsurugi K (1987) The mechanism of action of ricin and related toxic lectins on eukaryotic ribosomes. The site and the characteristics of the modification in 28 S ribosomal RNA caused by the toxins. J Biol Chem 262:5908–5912
Endo Y, Tsurugi K (1987) RNA N-glycosidase activity of ricin A chain. Mechanism of action of the toxic lectin ricin on eukaryotic ribosomes. J Biol Chem 262:8128–8130
Endo Y, Tsurugi K, Yutsudo T, Takeda Y, Ogasawara T, Igarashi K (1988) Site of action of a Vero toxin (VT2) from Escherichia coli O157:H7 and of Shiga toxin on eukaryotic ribosomes. RNA N-glycosidase activity of the toxins. Eur J Biochem 171:45–50
Ewers H, Romer W, Smith AE, Bacia K, Dmitrieff S, Chai W, Mancini R, Kartenbeck J, Chambon V, Berland L, Oppenheim A, Schwarzmann G, Feizi T, Schwille P, Sens P, Helenius A, Johannes L (2010) GM1 structure determines SV40-induced membrane invagination and infection. Nat Cell Biol 12:11–18; sup pp 1–12
Falguieres T, Johannes L (2006) Shiga toxin B-subunit binds to the chaperone BiP and the nucleolar protein B23. Biol Cell 98:125–134
Feng Y, Jadhav AP, Rodighiero C, Fujinaga Y, Kirchhausen T, Lencer WI (2004) Retrograde transport of cholera toxin from the plasma membrane to the endoplasmic reticulum requires the trans-Golgi network but not the Golgi apparatus in Exo2-treated cells. EMBO Rep 5:596–601
Ferrari DM, Söling HD (1999) The protein disulphide-isomerase family: unravelling a string of folds. Biochem J 339(Pt 1):1–10
Forster ML, Sivick K, Park YN, Arvan P, Lencer WI, Tsai B (2006) Protein disulfide isomerase-like proteins play opposing roles during retrotranslocation. J Cell Biol 173:853–859
Fraser ME, Fujinaga M, Cherney MM, Melton-Celsa AR, Twiddy EM, O’Brien AD, James MN (2004) Structure of shiga toxin type 2 (Stx2) from Escherichia coli O157:H7. J Biol Chem 279:27511–27517
Gardner RG, Swarbrick GM, Bays NW, Cronin SR, Wilhovsky S, Seelig L, Kim C, Hampton RY (2000) Endoplasmic reticulum degradation requires lumen to cytosol signaling. Transmembrane control of Hrd1p by Hrd3p. J Cell Biol 151:69–82
Garred O, van Deurs B, Sandvig K (1995) Furin-induced cleavage and activation of Shiga toxin. J Biol Chem 270:10817–10821
Gauss R, Jarosch E, Sommer T, Hirsch C (2006) A complex of Yos9p and the HRD ligase integrates endoplasmic reticulum quality control into the degradation machinery. Nat Cell Biol 8:849–854
Giodini A, Cresswell P (2008) Hsp90-mediated cytosolic refolding of exogenous proteins internalized by dendritic cells. EMBO J 27:201–211
Girard M, Poupon V, Blondeau F, McPherson PS (2005) The DnaJ-domain protein RME-8 functions in endosomal trafficking. J Biol Chem 280:40135–40143
Guetzoyan LJ, Spooner RA, Boal F, Stephens DJ, Lord JM, Roberts LM, Clarkson GJ (2010) Fine tuning Exo2, a small molecule inhibitor of secretion and retrograde trafficking pathways in mammalian cells. Mol Biosyst 6:2030–2038
Harley SM, Beevers H (1982) Ricin inhibition of in vitro protein synthesis by plant ribosomes. Proc Natl Acad Sci U S A 79:5935–5938
Harley SM, Beevers H (1986) Lectins in castor bean seedlings. Plant Physiol 80:1–6
Hazes B, Read RJ (1997) Accumulating evidence suggests that several AB-toxins subvert the endoplasmic reticulum-associated protein degradation pathway to enter target cells. Biochemistry 36:11051–11054
Heiligenstein S, Eisfeld K, Sendzik T, Jimenez-Becker N, Breinig F, Schmitt MJ (2006) Retrotranslocation of a viral A/B toxin from the yeast endoplasmic reticulum is independent of ubiquitination and ERAD. EMBO J 25:4717–4727
Heyningen SV (1974) Cholera toxin: interaction of subunits with ganglioside GM1. Science 183:656–657
Jacewicz M, Feldman HA, Donohue-Rolfe A, Balasubramanian KA, Keusch GT (1989) Pathogenesis of Shigella diarrhea XIV. Analysis of Shiga toxin receptors on cloned HeLa cells. J Infect Dis 159:881–889
Jakob CA, Burda P, Roth J, Aebi M (1998) Degradation of misfolded endoplasmic reticulum glycoproteins in Saccharomyces cerevisiae is determined by a specific oligosaccharide structure. J Cell Biol 142:1223–1233
Jarosch E, Taxis C, Volkwein C, Bordallo J, Finley D, Wolf DH, Sommer T (2002) Protein dislocation from the ER requires polyubiquitination and the AAA-ATPase Cdc48. Nat Cell Biol 4:134–139
Johannes L, Decaudin D (2005) Protein toxins: intracellular trafficking for targeted therapy. Gene Ther 12:1360–1368
Johannes L, Popoff V (2008) Tracing the retrograde route in protein trafficking. Cell 135:1175–1187
Kanehara K, Kawaguchi S, Ng DT (2007) The EDEM and Yos9p families of lectin-like ERAD factors. Semin Cell Dev Biol 18:743–750
Kanehara K, Xie W, Ng DT (2010) Modularity of the Hrd1 ERAD complex underlies its diverse client range. J Cell Biol 188:707–716
Klappa P, Hawkins HC, Freedman RB (1997) Interactions between protein disulphide isomerase and peptides. Eur J Biochem 248:37–42
Kothe M, Ye Y, Wagner JS, De Luca HE, Kern E, Rapoport TA, Lencer WI (2005) Role of p97 AAA-ATPase in the retrotranslocation of the cholera toxin A1 chain, a non-ubiquitinated substrate. J Biol Chem 280:28127–28132
LaPointe P, Wei X, Gariepy J (2005) A role for the protease-sensitive loop region of Shiga-like toxin 1 in the retrotranslocation of its A1 domain from the endoplasmic reticulum lumen. J Biol Chem 280:23310–23318
Li S, Spooner RA, Allen SC, Guise CP, Ladds G, Schnoder T, Schmitt MJ, Lord JM, Roberts LM (2010) Folding-competent and folding-defective forms of ricin A chain have different fates after retrotranslocation from the endoplasmic reticulum. Mol Biol Cell 21:2543–2554
Lipson C, Alalouf G, Bajorek M, Rabinovich E, Atir-Lande A, Glickman M, Bar-Nun S (2008) A proteasomal ATPase contributes to dislocation of endoplasmic reticulum-associated degradation (ERAD) substrates. J Biol Chem 283:7166–7175
Majoul I, Ferrari D, Söling HD (1997) Reduction of protein disulfide bonds in an oxidizing environment. The disulfide bridge of cholera toxin A-subunit is reduced in the endoplasmic reticulum. FEBS Lett 401:104–108
Majoul IV, Bastiaens PI, Soling HD (1996) Transport of an external Lys-Asp-Glu-Leu (KDEL) protein from the plasma membrane to the endoplasmic reticulum: studies with cholera toxin in Vero cells. J Cell Biol 133:777–789
Marshall RS, Jolliffe NA, Ceriotti A, Snowden CJ, Lord JM, Frigerio L, Roberts LM (2008) The role of CDC48 in the retro-translocation of non-ubiquitinated toxin substrates in plant cells. J Biol Chem 283:15869–15877
Massey S, Banerjee T, Pande AH, Taylor M, Tatulian SA, Teter K (2009) Stabilization of the tertiary structure of the cholera toxin A1 subunit inhibits toxin dislocation and cellular intoxication. J Mol Biol 393:1083–1096
Mayerhofer PU, Cook JP, Wahlman J, Pinheiro TT, Moore KA, Lord JM, Johnson AE, Roberts LM (2009) Ricin A chain insertion into endoplasmic reticulum membranes is triggered by a temperature increase to 37°C. J Biol Chem 284:10232–10242
McConnell E, Lass A, Wojcik C (2007) Ufd1-Npl4 is a negative regulator of cholera toxin retrotranslocation. Biochem Biophys Res Commun 355:1087–1090
McKenzie J, Johannes L, Taguchi T, Sheff D (2009) Passage through the Golgi is necessary for Shiga toxin B subunit to reach the endoplasmic reticulum. Febs J 276:1581–1595
Menikh A, Saleh MT, Gariepy J, Boggs JM (1997) Orientation in lipid bilayers of a synthetic peptide representing the C-terminus of the A1 domain of shiga toxin. A polarized ATR-FTIR study. Biochemistry 36:15865–15872
Nichols BJ, Kenworthy AK, Polishchuk RS, Lodge R, Roberts TH, Hirschberg K, Phair RD, Lippincott-Schwartz J (2001) Rapid cycling of lipid raft markers between the cell surface and Golgi complex. J Cell Biol 153:529–541
O’Neal CJ, Amaya EI, Jobling MG, Holmes RK, Hol WG (2004) Crystal structures of an intrinsically active cholera toxin mutant yield insight into the toxin activation mechanism. Biochemistry 43:3772–3782
Olsnes S, Saltvedt E, Pihl A (1974) Isolation and comparison of galactose-binding lectins from Abrus precatorius and Ricinus communis. J Biol Chem 249:803–810
Orlandi PA (1997) Protein-disulfide isomerase-mediated reduction of the A subunit of cholera toxin in a human intestinal cell line. J Biol Chem 272:4591–4599
Pande AH, Scaglione P, Taylor M, Nemec KN, Tuthill S, Moe D, Holmes RK, Tatulian SA, Teter K (2007) Conformational instability of the cholera toxin A1 polypeptide. J Mol Biol 374:1114–1128
Pawar V, De A, Briggs L, Omar MM, Sweeney ST, Lord JM, Roberts LM, Spooner RA, Moffat KG (2011) RNAi screening of Drosophila (Sophophora) melanogaster S2 cells for Ricin sensitivity and resistance. J Biomol Screen 16:436–442
Popoff V, Mardones GA, Bai SK, Chambon V, Tenza D, Burgos PV, Shi A, Benaroch P, Urbe S, Lamaze C, Grant BD, Raposo G, Johannes L (2009) Analysis of articulation between clathrin and retromer in retrograde sorting on early endosomes. Traffic 10:1868–1880
Rabinovich E, Kerem A, Frohlich KU, Diamant N, Bar-Nun S (2002) AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol Cell Biol 22:626–634
Rapak A, Falnes PO, Olsnes S (1997) Retrograde transport of mutant ricin to the endoplasmic reticulum with subsequent translocation to cytosol. Proc Natl Acad Sci U S A 94:3783–3788
Reeves PJ, Callewaert N, Contreras R, Khorana HG (2002) Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc Natl Acad Sci U S A 99:13419–13424
Rodighiero C, Tsai B, Rapoport TA, Lencer WI (2002) Role of ubiquitination in retro-translocation of cholera toxin and escape of cytosolic degradation. EMBO Rep 3:1222–1227
Römer W, Berland L, Chambon V, Gaus K, Windschiegl B, Tenza D, Aly MR, Fraisier V, Florent JC, Perrais D, Lamaze C, Raposo G, Steinem C, Sens P, Bassereau P, Johannes L (2007) Shiga toxin induces tubular membrane invaginations for its uptake into cells. Nature 450:670–675
Rutenber E, Katzin BJ, Ernst S, Collins EJ, Mlsna D, Ready MP, Robertus JD (1991) Crystallographic refinement of ricin to 2.5 A. Proteins 10:240–250
Saleh MT, Ferguson J, Boggs JM, Gariepy J (1996) Insertion and orientation of a synthetic peptide representing the C-terminus of the A1 domain of Shiga toxin into phospholipid membranes. Biochemistry 35:9325–9334
Sandvig K, Ryd M, Garred O, Schweda E, Holm PK, van Deurs B (1994) Retrograde transport from the Golgi complex to the ER of both Shiga toxin and the nontoxic Shiga B-fragment is regulated by butyric acid and cAMP. J Cell Biol 126:53–64
Sandvig K, van Deurs B (2005) Delivery into cells: lessons learned from plant and bacterial toxins. Gene Ther 12:865–872
Saslowsky DE, Cho JA, Chinnapen H, Massol RH, Chinnapen DJ, Wagner JS, De Luca HE, Kam W, Paw BH, Lencer WI (2010) Intoxication of zebrafish and mammalian cells by cholera toxin depends on the flotillin/reggie proteins but not Derlin-1 or -2. J Clin Invest
Sharp GW, Hynie S (1971) Stimulation of intestinal adenyl cyclase by cholera toxin. Nature 229:266–269
Simpson JC, Dascher C, Roberts LM, Lord JM, Balch WE (1995a) Ricin cytotoxicity is sensitive to recycling between the endoplasmic reticulum and the Golgi complex. J Biol Chem 270:20078–20083
Simpson JC, Lord JM, Roberts LM (1995b) Point mutations in the hydrophobic C-terminal region of ricin A chain indicate that Pro250 plays a key role in membrane translocation. Eur J Biochem 232:458–463
Simpson JC, Roberts LM, Lord JM (1995c) Catalytic and cytotoxic activities of recombinant ricin A chain mutants with charged residues added at the carboxyl terminus. Protein Expr Purif 6:665–670
Simpson JC, Roberts LM, Romisch K, Davey J, Wolf DH, Lord JM (1999) Ricin A chain utilises the endoplasmic reticulum-associated protein degradation pathway to enter the cytosol of yeast. FEBS Lett 459:80–84
Slominska-Wojewodzka M, Gregers TF, Walchli S, Sandvig K (2006) EDEM is involved in retrotranslocation of ricin from the endoplasmic reticulum to the cytosol. Mol Biol Cell 17:1664–1675
Spear ED, Ng DT (2005) Single, context-specific glycans can target misfolded glycoproteins for ER-associated degradation. J Cell Biol 169:73–82
Spilsberg B, Van Meer G, Sandvig K (2003) Role of lipids in the retrograde pathway of ricin intoxication. Traffic 4:544–552
Spooner RA, Hart PJ, Cook JP, Pietroni P, Rogon C, Hohfeld J, Roberts LM, Lord JM (2008a) Cytosolic chaperones influence the fate of a toxin dislocated from the endoplasmic reticulum. Proc Natl Acad Sci U S A 105:17408–17413
Spooner RA, Smith DC, Easton AJ, Roberts LM, Lord JM (2006) Retrograde transport pathways utilised by viruses and protein toxins. Virol J 3:26
Spooner RA, Watson P (2010) Drug targeting: learning from toxin entry and trafficking in mammalian cells. Curr Opin Drug Discov Devel 13:86–95
Spooner RA, Watson P, Smith DC, Boal F, Amessou M, Johannes L, Clarkson GJ, Lord JM, Stephens DJ, Roberts LM (2008b) The secretion inhibitor Exo2 perturbs trafficking of Shiga toxin between endosomes and the trans-Golgi network. Biochem J 414:471–484
Spooner RA, Watson PD, Marsden CJ, Smith DC, Moore KA, Cook JP, Lord JM, Roberts LM (2004) Protein disulphide-isomerase reduces ricin to its A and B chains in the endoplasmic reticulum. Biochem J 383:285–293
Surjit M, Jameel S, Lal SK (2007) Cytoplasmic localization of the ORF2 protein of hepatitis E virus is dependent on its ability to undergo retrotranslocation from the endoplasmic reticulum. J Virol 81:3339–3345
Tam PJ, Lingwood CA (2007) Membrane cytosolic translocation of verotoxin A1 subunit in target cells. Microbiology 153:2700–2710
Taylor M, Navarro-Garcia F, Huerta J, Burress H, Massey S, Ireton K, Teter K (2010) Hsp90 is required for transfer of the cholera toxin A1 subunit from the endoplasmic reticulum to the cytosol. J Biol Chem 285:31261–31267
Tsai B, Rodighiero C, Lencer WI, Rapoport TA (2001) Protein disulfide isomerase acts as a redox-dependent chaperone to unfold cholera toxin. Cell 104:937–948
Ushioda R, Hoseki J, Araki K, Jansen G, Thomas DY, Nagata K (2008) ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science 321:569–572
Valeva A, Walev I, Weis S, Boukhallouk F, Wassenaar TM, Endres K, Fahrenholz F, Bhakdi S, Zitzer A (2004) A cellular metalloproteinase activates Vibrio cholerae pro-cytolysin. J Biol Chem 279:25143–25148
Vembar SS, Brodsky JL (2008) One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol 9:944–957
Wales R, Chaddock JA, Roberts LM, Lord JM (1992) Addition of an ER retention signal to the ricin A chain increases the cytotoxicity of the holotoxin. Exp Cell Res 203:1–4
Wernick NL, De Luca H, Kam WR, Lencer WI (2010) N-terminal extension of the cholera toxin A1-chain causes rapid degradation after retrotranslocation from endoplasmic reticulum to cytosol. J Biol Chem 285:6145–6152
Wesche J, Rapak A, Olsnes S (1999) Dependence of ricin toxicity on translocation of the toxin A-chain from the endoplasmic reticulum to the cytosol. J Biol Chem 274:34443–34449
Windschiegl B, Orth A, Römer W, Berland L, Stechmann B, Bassereau P, Johannes L, Steinem C (2009) Lipid reorganization induced by Shiga toxin clustering on planar membranes. PLoS One 4:e6238
Winkeler A, Godderz D, Herzog V, Schmitz A (2003) BiP-dependent export of cholera toxin from endoplasmic reticulum-derived microsomes. FEBS Lett 554:439–442
Xie W, Kanehara K, Sayeed A, Ng DT (2009) Intrinsic conformational determinants signal protein misfolding to the Hrd1/Htm1 endoplasmic reticulum-associated degradation system. Mol Biol Cell 20:3317–3329
Ye Y, Meyer HH, Rapoport TA (2001) The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature 414:652–656
Yu M, Haslam DB (2005) Shiga toxin is transported from the endoplasmic reticulum following interaction with the luminal chaperone HEDJ/ERdj3. Infect Immun 73:2524–2532
Acknowledgments
This work was supported by Wellcome Trust Programme Grant 080566/Z/06/Z and National Institutes of Health Grant 5U01AI65869-02.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2011 Springer-Verlag Berlin Heidelberg
About this chapter
Cite this chapter
Spooner, R.A., Lord, J.M. (2011). How Ricin and Shiga Toxin Reach the Cytosol of Target Cells: Retrotranslocation from the Endoplasmic Reticulum. In: Mantis, N. (eds) Ricin and Shiga Toxins. Current Topics in Microbiology and Immunology, vol 357. Springer, Berlin, Heidelberg. https://doi.org/10.1007/82_2011_154
Download citation
DOI: https://doi.org/10.1007/82_2011_154
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-27469-5
Online ISBN: 978-3-642-27470-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)