Abstract
We describe the first genome isolation of Middle East respiratory syndrome coronavirus (MERS-CoV) in Kenya. This fatal zoonotic pathogen was first described in the Kingdom of Saudi Arabia in 2012. Epidemiological and molecular evidence revealed zoonotic transmission from camels to humans and between humans. Currently, MERS-CoV is classified by the WHO as having high pandemic potential requiring greater surveillance. Previous studies of MERS-CoV in Kenya mainly focused on site-specific and archived camel and human serum samples for antibodies. We conducted active nationwide cross-sectional surveillance of camels and humans in Kenya, targeting both nasal swabs and plasma samples from 1,163 camels and 486 humans collected from January 2016 to June 2018. A total of 792 camel plasma samples were positive by ELISA. Seroprevalence increased with age, and the highest prevalence was observed in adult camels (82.37%, 95% confidence interval (CI) 79.50–84.91). More female camels were significantly seropositive (74.28%, 95% CI 71.14–77.19) than male camels (P < 0.001) (53.74%, 95% CI 48.48–58.90). Only 11 camel nasal swabs were positive for MERS-CoV by reverse transcription-quantitative PCR. Phylogenetic analysis of whole genome sequences showed that Kenyan MERS-CoV clustered within sub-clade C2, which is associated with the African clade, but did not contain signature deletions of orf4b in African viruses. None of the human plasma screened contained neutralizing antibodies against MERS-CoV. This study confirms the geographically widespread occurrence of MERS-CoV in Kenyan camels. Further one-health surveillance approaches in camels, wildlife, and human populations are needed.
Similar content being viewed by others
Introduction
Middle East respiratory syndrome coronavirus (MERS-CoV) is a positive sense, single-stranded RNA virus in the genus Betacoronavirus. It is a zoonotic pathogen capable of causing severe respiratory disease in humans. Dromedary camels are considered as a source of zoonotic origin and natural reservoir host for MERS-CoV (Azhar et al.2014; Haagmans et al.2014). As of September 20, 2018, MERS-CoV infection has been reported from 27 countries with 2249 laboratory-confirmed cases in humans and at least 798 related deaths (WHO 2018). Most of these cases occurred in the Kingdom of Saudi Arabia, where the high prevalence of MERS-CoV in dromedary camels and direct contact with infected camels have been linked to human infections (Haagmans et al.2014; Azhar et al.2014). Notably, most human cases outside of the Arabian Peninsula have been linked to travel from Saudi Arabia. Numerous surveillance studies in Africa have revealed the presence of MERS-CoV antibodies in dromedaries from several African countries including Nigeria, Egypt, and Mali (Chu et al.2014; Chu et al.2018; Chu et al.2015; Falzarano et al.2017), where camels are reared and people frequently travel for pilgrimage to Saudi Arabia.
Similarly, in Kenya, previous studies reported a high seroprevalence of MERS-CoV antibodies in archived dromedaries samples collected in 1992–2013 (Corman et al.2014), and serological infection of MERS-CoV was confirmed in two people sampled from Tana River County in 2013–2014 (Liljander et al.2016). However, a recent study reported no evidence of MERS-CoV infection among camel farmers (Munyua et al.2017). Other recent studies suggested that the high densities of camel populations may be correlated with increased seropositivity and may contribute to long-term maintenance of the virus in the camel population. In this study, we evaluated the exposure of camels and humans to MERS-CoV in all camel-rearing counties in Kenya as part of a country-wide surveillance program.
Materials and Methods
Study Area and Design
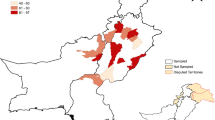
This cross-sectional study was carried out from January 2016 to June 2018 across 13 counties in Kenya where camels are reared (Table 1 and Fig. 1). The four camel breed types in Kenya sampled in this study were from historical and recent camel rearing tribes (Mburu et al.2003). Therefore, based on the geographical division of breed types, the 13 counties were grouped into five groups. For human sampling, high-risk groups such as camel herders and their immediate families were targeted. Sampling mainly focused on high-risk locations such as around watering points and common browsing locations that attract several herds of camels from different regions.
Sampling Procedure
For camel sampling, camel owners were informed about the study in a language that they understood. The camels were physically restrained, and 10 mL of blood from the jugular vein was drawn into anti-coagulant vacuum blood collection tubes. Additionally, 1163 nasal swabs samples were collected in RNAlater® (Ambion, Foster City, CA, USA) and virus transport medium.
The research was conducted in accordance with the Helsinki Declaration for sampling of human subjects. Informed consent was obtained from camel owners, handlers, and family members or their guardians (in case of underage children) from whom blood was collected. Five milliliters of blood from consenting study participants were collected by a clinician or phlebotomist from a peripheral vein into an anti-coagulant vacuum blood collection tube. Sociodemographic data were also collected from the participants (Supplemental Table S2). Other data such as age and sex were also collected.
Blood samples were centrifuged, plasma was aliquoted and stored in liquid nitrogen while in the field, and the samples were transported to Nairobi for storage at − 80 °C. A total of 1,163 dromedary camels were sampled from 13 counties and 486 human subjects from 10 counties. Camel and human samples were exported to Wuhan Institute of Virology, Chinese Academy of Science in Hubei, Peoples Republic of China. All samples were transported according to IATA international regulations for transporting viable samples.
Serology Testing
An in-house anti-MERS-CoV IgG ELISA kit was developed based on the purified spike protein receptor binding domain. This highly sensitive and specific ELISA was previously validated for use with samples from camel and human (Zohaib et al.2018). This anti-MERS-CoV IgG ELISA was used with minor modifications. Camel samples were tested at 1:20 dilution and goat anti-camel IgG-horseradish peroxidase conjugate (Alpha Diagnostic International, San Antonio, TX, USA) was used as the secondary antibody at 1:3000. Based on the microneutralization test, a cut-off value of 0.35 was determined. For human samples, plasma was tested at a dilution of 1:20 and anti-human IgG-horseradish peroxidase conjugated monoclonal antibody (Kyab Biotech Co., Ltd, Wuhan, China) was used as the secondary antibody at 1:15000.
Microneutralization Assay
A microneutralization assay was performed as described previously (Perera et al. 2013). Briefly, Vero B4 cells were seeded into 96-well plates. Plasma samples were incubated at 56 °C for 1 h. MERS-CoV (EMC strain) was diluted with DMEM to 100 TCID50/50 μL. The plasma samples were diluted by twofold in DMEM and incubated with MERS-CoV at 37 °C for 30 min. The medium was removed from the cells and 50 μL virus-plasma mixture was added. The virus-plasma mixture was removed after 1 h and 100 μL DMEM plus 2% fetal bovine serum (FBS) and 1% penicillin/streptomycin was added. The cells were incubated at 37 °C with 5% CO2, and the cytopathic effect (CPE) was observed and recorded at 4 days post-infection. Samples that inhibited CPE at a dilution of 1:20 were considered as positive.
Molecular Detection of MERS-CoV in Camel Nasal Swabs
Viral RNA was extracted from camel nasal swabs stored in RNAlater® (Ambion) using a viral RNA extraction kit (Roche, Basel, Switzerland) according to the manufacturer’s instructions. All camel nasal swabs were screened for MERS-CoV using two independent TaqMan quantitative reverse transcription PCR assays for the nucleocapsid gene (N) according to the WHO testing algorithm as described previously (Lu et al.2014). Additionally, camel samples positive according to RT-qPCR were also screened by MERS-CoV-specific RT-PCR targeting the N gene to confirm the presence of MERS-CoV in the samples as described previously (Corman et al.2012).
MERS-CoV Isolation
Virus isolation from two positive nasal swabs with high viral loads was attempted using Vero cells. Vero cell monolayers were maintained in DMEM supplemented with 10% FBS. Nasal swab specimens in viral transport medium were diluted in DMEM before being added to the Vero cells. After incubation at 37 °C for 1.5 h, the inoculum was removed and replaced with fresh DMEM containing 2% FBS and antibiotics. The cells were incubated at 37 °C for 5 days and observed daily for CPEs. The culture supernatant and cells were examined for the presence of virus by the RT-qPCR N2 assay targeting the MERS-CoV N gene (Corman et al. 2012) and immunofluorescence assay using a rabbit antibody (prepared in house) against the MERS-CoV N protein.
Full Genome Sequencing of MERS-CoV and Analysis
Libraries for next-generation sequencing were prepared using an Illumina Truseq mRNA kit (TruSeq Stranded mRNA Library Prep Kit, Cat #RS-122-2101, Illumina, San Diego, CA, USA) following the manufacturer’s instructions. The sequencing was performed on a HiSeq 3000 sequencer. The data obtained was analyzed in Metavisitor (a suite of galaxy tools) as described previously (Carissimo et al. 2017). The BLAST-guided scaffold was then used to reference align the reads in Geneious R11. Phylogenetic analysis was performed in MEGA7. For MERS-CoV samples that were not selected for full-genome sequencing, specific RT-PCRs were set-up to amplify the partial S gene and fragment covering the gene regions of orf3, orf4a, and orf4b as described previously (Smits et al.2015; Chu et al.2018).
Statistical Analysis
ELISA-positive samples were statistically analyzed (Chi Square test) to detect associations between location, age, and sex. Univariable analysis was performed and odds ratios along with their 95% confidence intervals (CIs) were calculated. A P value < 0.05 was considered as significant in all analyses. Statistical analysis was performed in R (v3.5.1) with epicalc (v2.15.1.0) and the DescTools (v0.99.25) packages.
Results
MERS-CoV Seroprevalence among Camel Populations in Kenya
A total of 1163 plasma samples was collected from camels in 13 counties in Kenya between January 2016 to June 2018. Age and gender data for the 14 camels were missing. From the remaining 1149 samples, 801 (69.71%) were female and 348 (30.29%) were male. Most plasma samples (611; 52.54%) were collected from the northeastern part of Kenya (region C), which also has the largest herd of camels in Kenya and borders the Republic of Somalia. The demographic distribution of plasma in different camel breed regions and administrative counties is presented in Table 1 and Fig. 1. A total of 792 of the 1163 (68.10%) camel plasma samples tested positive by ELISA. Seroprevalence varied significantly (P < 0.001) among regions in the country, ranging from the highest in region B (79.86, 95% CI 74.90–84.06) followed by region C (75.29, 95% CI 71.72–78.54), region A (48.72, 95% CI 41.00–56.50), region E (42.11%, 95% CI 23.14–63.72), and region D (16.67%, 95% CI 10.20–26.05). The significantly (P < 0.001) highest prevalence was observed in Marsabit county (87.34%, 95% CI 78.24–92.98) (Supplemental Table S1). The seroprevalence of MERS-CoV increased with age and was significantly higher (P < 0.001) in adult camels > 7 years (82.37%, 95% CI 79.50–84.91) compared to sub-adults > 4 years < 7 years (58.57%, 95% CI 46.88–69.37) and juvenile camels < 4 years (36.05%, 95% CI 30.98–41.46). Significantly higher (P < 0.001) seroprevalence was observed in female (74.28%, 95% CI 71.14–77.19) than in male camels (53.74%, 95% CI 48.48–58.90). The geographical location and age of the camels were the main factors affecting the MERS-CoV seroprevalence in Kenya (Table 1).
MERS-CoV Seroprevalence in Humans in Kenya
A total of 486 human plasma samples were collected from 10 counties in Kenya from March 2017 to June 2018, among which 231 (48.63%) subjects were male and 244 (51.37%) were female; gender data for 11 human samples were missing. Among the humans sampled, 95.27% stated that they have had close contact with livestock including camels. Three hundred people in this group stated they have regular contact with camels as herdsmen.
Twenty of the 486 human plasma samples showed positive results by ELISA. Of these, eight were from West Pokot, five were from Tana River, four were from Garissa, two were from Wajir, and one was from Isiolo Counties. Ten seropositive samples were male and nine were female, gender data of one ELISA reactive sample was missing. Twelve of the 20 ELISA-positive individuals were in frequent contact with camels. A micro-neutralization test was performed on all ELISA-reactive samples and none were positive in the neutralization assay.
Molecular Detection of MERS-CoV
Eleven camel nasal swabs (Table 2) were positive for both N2 and N3 by RT-qPCR and nested PCR for the N gene. Among them, five were from adult camels, four were from juvenile camels, and one was a sub-adult camel. Age data for one positive camel was not available. High viral loads were observed in juvenile camels compared to in adults. Sequencing of the PCR-positive N gene samples revealed 100% identical sequences. Contamination was ruled out by repeating the experiments in independent laboratories and targeted PCR amplification and sequencing of the N gene. We partially sequenced the S gene of eight samples as described previously (Table 2) (Smits et al.2015). Of these eight, six samples were 100% identical, whereas the other two samples showed one nucleotide difference compared to the other six positive samples. All MERS-CoV isolates from Kenya clusters within sub-clade C2, which is associated with the African clade (data not shown).
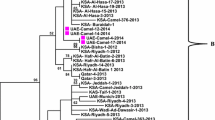
Of the 11 samples detected in this study, two specimens with a high viral load were selected for virus isolation and full-length sequencing. Cytopathic changes in Vero cells were observed sequentially for 5 days post-infection (Fig. 2A). Camel MERS-CoV isolates in Vero cells were confirmed by immunofluorescence assay (Fig. 2B) and RT-qPCR (data not shown). Genetic nucleotide identity was 100% between two Kenyan viruses and 99.63%–99.77% for viruses from sub-clade C2 (Fig. 3), > 99.14% within camel and human MERS-CoV from the Middle East, and 97.82%–98.10% for viruses from sub-clade C1. Full-length sequencing was also performed on RNAlater® preserved nasal swab samples. No nucleic acid differences were detected between isolated viruses and RNAlater® preserved samples. The results of phylogenetic analysis of Kenyan MERS-CoV sequences along with other relevant viruses are shown in Fig. 3. The tree was rooted against a MERS-CoV-related bat coronavirus from South Africa (KC869678.4). Viruses from Kenya clustered together with sequences from Ethiopia, Egypt, Burkina Faso, Nigeria, and Morocco in clade C. Within clade C, viruses from Kenya clustered with viruses from Ethiopia and Egypt in sub-clade C2. Characteristics signature deletions in orf4b have been observed in viruses from Africa in sub-clade C1 but not in those from Egypt and sub-clade C2. Notably, Ethiopian and Egyptian MERS-CoV encode full-length orf4b (246 amino acids). Two viruses detected in this study were unique because they had a truncated orf4b (244aa) in sub-clade C2 (Fig. 4), whereas the other three sequences encoded full-length orf4b of 246 amino acids. Previously, a deletion pattern in the orf3 region of African MERS-CoV has also been reported (Chu et al.2018). However, MERS-CoV from Kenya encodes full-length orf3. The amino acid residues of Kenyan sample C1215 differed from the EMC strain throughout the virus genome. The amino acid residues in the spike protein receptor binding domain of C1215 contained the amino acid substitutions S10F and S148P in the receptor binding motif. Although these two mutations were observed in the spike receptor binding domain region of MERS-CoV, the camel plasma and human plasma were effectively neutralized by the EMC strain from Kenya as described previously; thus these mutations likely do not affect the affinity of the protein for the host receptor (Corman et al.2014; Liljander et al.2016), although further studies are needed to confirm this.
Isolation of camel MERS-CoV C1215 and C1272. A Induction of cytopathic effect on Vero cells. The images were taken by NIS Elements F (ECLIPSE TS100, Nikon). Original magnification: 100 ×. B Successful isolation of camel MERS-CoV was confirmed by immunofluorescent antibody staining using rabbit antibody against the MERS-CoV N protein. The columns (from left to right) show staining of nuclei (blue), virus replication (red), and both nuclei and virus replication (merged double-stain images). The images were taken by a confocal microscope. Scale bar = 100 μm.
Phylogenetic analysis of MERS-CoV full genomes using neighbor-joining method in MEGA7. Bootstrap values of nodes are shown. Bootstrap values along branches are for 1,000 replicates. The tree was rooted against a MERS-CoV related bat coronavirus Neoromicia/PML-PHE1/RSA/2011 (KC869678) from South Africa. To allow for greater resolution of the viruses of interest, the long branch of KC869678 was removed. Detected MERS-CoV viruses in this study are red colored and identified with circle node markers (●). Scale bar indicates nucleotide substitutions per site.
Discussion
Previous studies have reported the prevalence of MERS-CoV antibodies in archived samples (Corman et al.2014; Liljander et al.2016; Munyua et al.2017). This is the first study to report the detection and characterization of MERS-CoV from Kenyan dromedaries. The nucleic acid sequences determined in this study confirm the genetic uniqueness of the African clade of this virus compared to those reported in the Arabian Peninsula (Chu et al.2018). The observed genetic relationship between MERS-CoV sequences in Kenya and those detected in camels from Ethiopia are likely related to strong geographical affinities. A recent study characterized MERS-CoV from Africa, which exhibited region-dependent genetic diversity (Chu et al.2018). However, the study did not describe MERS-CoV from Kenya. In the current study, we found that MERS-CoV from Kenya is phylogenetically distinct (Clade C) from viruses in the Arabian Peninsula. Additionally, although truncation of orf4b was observed in Kenyan viruses, these viruses lacked the signature deletion patterns of orf4b observed previously in western African MERS-CoV in sub-clade C1. Previous studies focused on genetic and phenotypic characterization of MERS-CoV from West Africa; however, studies of Eastern African MERS-CoV utilizing one-health approaches are urgently needed.
This study further confirms the widespread presence of MERS-CoV among camels in all camel-rearing counties in Kenya. Although the seroprevalence varied between counties in Kenya, the widespread occurrence indicates the need for a national-wide strategy for early detection and prevention rather than county-based approaches. The seroprevalence rates observed in this study are consistent with those obtained in previous studies in Kenya and other African countries. Of the 13 counties evaluated in this study, the highest seropositivity was observed in Marsabit county (87.34%), which is comparable to that reported in a recent study of archived plasma of Marsabit county camels in 2013.
Eleven camels were positive for MERS-CoV nucleic acids in different counties, indicating the active circulation of MERS-CoV in Kenya, which was confirmed by the presence of MERS-CoV antibodies in juvenile/young camels in all geographic regions except for in region E. This agrees with the results of previous studies which suggested that younger camels are most likely to be infected by MERS-CoV (Sabir et al.2016). The presence of a high viral load in juvenile camels also suggests that younger camels play a key role in the maintenance and spread of MERS-CoV.
We were not able to sample an equal proportion of males and females mainly because of camel husbandry practices which value female camels, which produce milk, over males. Our study focused on high-risk groups i.e. humans that frequently interact with camels as well as infection hot spots such as common watering points and browsing locations. Most herders prefer moving camels in several groups for feeding and watering as a form of security. Additionally, watering points are frequented by different types of livestock such as cattle, sheep, and goats. Humans and wildlife also utilize these water sources, as these counties are in either arid or semi-arid regions where water is a scarce resource.
As previously reported, we were not able to detect neutralizing antibodies from sampled humans even though some of them were ELISA positive (Munyua et al.2017). We predict that MERS-CoV in Kenyan camels has low pathogenicity in humans, and neutralization antibody levels either decreased quickly or were undetectable in the neutralization assay; second, other coronaviruses are closely related to MERS-CoV but distinct at the virus neutralization epitopes. Additional studies focusing on surveillance in human, other wildlife species, and camels are needed to determine the prevalence of MERS-CoV in the general Kenyan population and identify the risk factors of infection.
Change history
28 February 2019
The acknowledgement section in the original article was published incorrectly.
References
Azhar EI, El-Kafrawy SA, Farraj SA, Hassan AM, Al-Saeed MS, Hashem AM, Madani TA (2014) Evidence for camel-to-human transmission of MERS coronavirus. N Engl J Med 370:2499–2505
Carissimo G, van den Beek M, Vernick KD, Antoniewski C (2017) Metavisitor, a suite of galaxy tools for simple and rapid detection and discovery of viruses in deep sequence data. PLoS ONE 12:e0168397
Chu DKW, Poon LLM, Gomaa MM, Shehata MM, Perera RAPM, Abu Zeid D, El Rifay AS, Siu LY, Guan Y, Webby RJ, Ali MA, Peiris M, Kayali G (2014) MERS coronaviruses in dromedary camels, Egypt. Emerg Infect Dis 20:1049–1053
Chu DKW, Oladipo JO, Perera RAPM, Kuranga SA, Chan SMS, Poon LLM, Peiris M (2015) Middle East respiratory syndrome coronavirus (MERS-CoV) in dromedary camels in Nigeria, 2015. Euro Surveill. https://doi.org/10.2807/1560-7917.es.2015.20.49.30086
Chu DKW, Hui KPY, Perera RAPM, Miguel E, Niemeyer D, Zhao J, Channappanavar R, Dudas G, Oladipo JO, Traoré A, Fassi-Fihri O, Ali A, Demissié GF, Muth D, Chan MCW, Nicholls JM, Meyerholz DK, Kuranga SA, Mamo G, Zhou Z, So RTY, Hemida MG, Webby RJ, Roger F, Rambaut A, Poon LLM, Perlman S, Drosten C, Chevalier V, Peiris M (2018) MERS coronaviruses from camels in Africa exhibit region-dependent genetic diversity. Proc Natl Acad Sci U S A 115:3144–3149
Corman VM, Müller MA, Costabel U, Timm J, Binger T, Meyer B, Kreher P, Lattwein E, Eschbach-Bludau M, Nitsche A, Bleicker T, Landt O, Schweiger B, Drexler JF, Osterhaus AD, Haagmans BL, Dittmer U, Bonin F, Wolff T, Drosten C (2012) Assays for laboratory confirmation of novel human coronavirus (hCoV-EMC) infections. Euro Surveill 17:20334
Corman VM, Jores J, Meyer B, Younan M, Liljander A, Said MY, Gluecks I, Lattwein E, Bosch B-J, Drexler JF, Bornstein S, Drosten C, Müller MA (2014) Antibodies against MERS coronavirus in dromedary camels, Kenya, 1992–2013. Emerg Infect Dis 20:1319–1322
Falzarano D, Kamissoko B, de Wit E, Maïga O, Cronin J, Samaké K, Traoré A, Milne-Price S, Munster VJ, Sogoba N, Niang M, Safronetz D, Feldmann H (2017) Dromedary camels in northern Mali have high seropositivity to MERS-CoV. One Health 3:41–43
Haagmans BL, Al Dhahiry SHS, Reusken CBEM, Raj VS, Galiano M, Myers R, Godeke G-J, Jonges M, Farag E, Diab A, Ghobashy H, Alhajri F, Al-Thani M, Al-Marri SA, Al Romaihi HE, Al Khal A, Bermingham A, Osterhaus ADME, AlHajri MM, Koopmans MPG (2014) Middle East respiratory syndrome coronavirus in dromedary camels: an outbreak investigation. Lancet Infect Dis 14:140–145
Liljander A, Meyer B, Jores J, Müller MA, Lattwein E, Njeru I, Bett B, Drosten C, Corman VM (2016) MERS-CoV antibodies in humans, Africa, 2013–2014. Emerg Infect Dis 22:1086–1089
Lu X, Whitaker B, Sakthivel SKK, Kamili S, Rose LE, Lowe L, Mohareb E, Elassal EM, Al-sanouri T, Haddadin A, Erdman DD (2014) Real-time reverse transcription-PCR assay panel for Middle East respiratory syndrome coronavirus. J Clin Microbiol 52:67–75
Mburu DN, Ochieng JW, Kuria SG, Jianlin H, Kaufmann B, Rege JEO, Hanotte O (2003) Genetic diversity and relationships of indigenous Kenyan camel (Camelus dromedarius) populations: implications for their classification. Anim Genet 34:26–32
Munyua P, Corman VM, Bitek A, Osoro E, Meyer B, Müller MA, Lattwein E, Thumbi SM, Murithi R, Widdowson M-A, Drosten C, Njenga MK (2017) No serologic evidence of middle east respiratory syndrome coronavirus infection among camel farmers exposed to highly seropositive camel herds: a household linked study, Kenya, 2013. Am J Trop Med Hyg 96:1318–1324
Perera RA, Wang P, Gomaa MR, El-Shesheny R, Kandeil A, Bagato O, Siu LY, Shehata MM, Kayed AS, Moatasim Y, Li M, Poon LL, Guan Y, Webby RJ, Ali MA, Peiris JS, Kayali G (2013) Seroepidemiology for MERS coronavirus using microneutralisation and pseudoparticle virus neutralisation assays reveal a high prevalence of antibody in dromedary camels in Egypt, June 2013. Eurosurveillance 18:20574
Sabir JSM, Lam TT-Y, Ahmed MMM, Li L, Shen Y, Abo-Aba SEM, Qureshi MI, Abu-Zeid M, Zhang Y, Khiyami MA, Alharbi NS, Hajrah NH, Sabir MJ, Mutwakil MHZ, Kabli SA, Alsulaimany FAS, Obaid AY, Zhou B, Smith DK, Holmes EC, Zhu H, Guan Y (2016) Co-circulation of three camel coronavirus species and recombination of MERS-CoVs in Saudi Arabia. Science 351:81–84
Smits SL, Raj VS, Pas SD, Reusken CBEM, Mohran K, Farag EABA, Al-Romaihi HE, AlHajri MM, Haagmans BL, Koopmans MP (2015) Reliable typing of MERS-CoV variants with a small genome fragment. J Clin Virol 64:83–87
World Health Organization (WHO) WHO | Middle East respiratory syndrome coronavirus (MERS-CoV). In: WHO. http://www.who.int/emergencies/mers-cov/en/. Accessed 29 May 2018
Zohaib A, Saqib M, Athar MA, Chen J, Sial A-R, Khan S, Taj Z, Halima S, Tahir U, Tayyab MH, Qureshi MA, Mansoor MK, Naeem MA, Hu B-J, Khan BA, Ujjan ID, Li B, Zhang W, Luo Y, Zhu Y, Waruhiu C, Khan I, Yang X-L, Sajid MS, Corman VM, Yan B, Shi Z-L (2018) Countrywide survey for MERS-Coronavirus antibodies in dromedaries and humans in Pakistan. Virol Sin 33:410–417
Acknowledgements
We thank all staff of the Ministry of Health and Ministry of Agriculture Livestock, Fisheries and Irrigation, Chiefs and local assistants from County governments of West Pokot, Turkana, Baringo, Samburu, Laikipia, Isiolo, Marsabit, Mandera, Wajir, Garissa, Tana River, Kitui, and Makueni; Kenya camel association; Kenya Wildlife Service; and National Museums of Kenya. Sheila Ommeh is a recipient of the Chinese Academy of Science Presidents International Fellowship Initiative (CAS-PIFI). This work was funded by Sino-Africa Joint Research Center (SAJC201313 and SAJC201605), External Cooperation Program of CAS (153211KYSB20160001), and National Science and Technology Major Project (2018ZX0101004).
Author information
Authors and Affiliations
Contributions
Z-LS and BA designed and supervised the overall study. SO, BH, X-YG, X-LY, MM, BA, and Z-LS coordinated the samplings. SO, BH, X-YG, X-LY, MM, VO, DO, and BA participated in sampling. SO, WZ, AZ, HZ, YL, SL, BH, JC, BL, CW, ZY, PZ, X-LY, L-FW, D-EA, and BY performed experiments. SO, WZ, AZ, BH, CW, ZY, PZ, L-FW, D-EA, BY, BA, and Z-LS analyzed data. SO, WZ, AZ, JC, BA and Z-LS wrote the manuscript. SO, WZ, AZ, BA, HZ, BH, X-YG, X-LY, MM, VO, YL, SL, CW, JC, BI, ZY, DO, VO, L-FW, D-EA, JL, EM, FG, PZ, K-JN, BY, and Z-LS interpreted data, revised and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Animal and Human Rights Statement
Permission for studying camels was granted by the Directorate of Veterinary Services at the State Department of Livestock, Ministry of Agriculture Livestock and Fisheries of Kenya, County veterinary departments, and study area chiefs. This study was approved by the Kenyatta National Hospital/University of Nairobi Ethics and Research Committee for conducting research on human subjects under permit reference number P210/04/2017. Informed consent was obtained from camel owners, handlers, and family members or their guardians (in case of underage children) from whom blood was collected. All institutional and National guidelines for care and handling use of animals were followed.
Open Access
This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Ommeh, S., Zhang, W., Zohaib, A. et al. Genetic Evidence of Middle East Respiratory Syndrome Coronavirus (MERS-Cov) and Widespread Seroprevalence among Camels in Kenya. Virol. Sin. 33, 484–492 (2018). https://doi.org/10.1007/s12250-018-0076-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12250-018-0076-4