Abstract
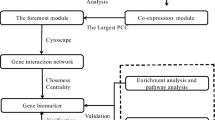
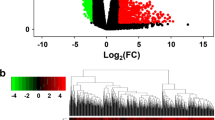
Hepatoblastoma (HB) is one of the most common liver malignancies in children, while the molecular basis of the disease is largely unknown. Therefore, this study aims to explore the key genes and molecular mechanisms of the pathogenesis of HB using a bioinformatics approach. The gene expression dataset GSE131329 was used to find differentially expressed genes (DEGs). Functional and enrichment analyses of the DEGs were performed by the EnrichR. Then, the protein-protein interaction (PPI) network of the up-regulated genes was constructed and visualized using STRING database and Cytoscape software, respectively. MCODE was used to detect the significant modules of the PPI network, and cytoHubba was utilized to rank the important nodes (genes) of the PPI modules. Overall, six ranking methods were employed and the results were validated by the Oncopression database. Moreover, the upstream regulatory network and the miRNA-target interactions of the up-regulated DEGs were analyzed by the X2K web and the miRTarBase respectively. A total of 594 DEGs, including 221 up- and 373 down-regulated genes, were obtained, which were enriched in different cellular and metabolic processes, human diseases, and cancer. Furthermore, 15 hub genes were screened, out of which, 11 were validated. Top 10 transcription factors, kinases, and miRNAs were also determined. To the best of our knowledge, the association of RACGAP1, MKI67, FOXM1, SIN3A, miR-193b, and miR-760 with HB was reported for the first time. Our findings may be used to shed light on the underlying mechanisms of HB and provide new insights for better prognosis and therapeutic strategies.




Similar content being viewed by others
References
Adesina AM, Lopez-Terrada D, Wong KK, Gunaratne P, Nguyen Y, Pulliam J, Margolin J, Finegold MJ (2009) Gene expression profiling reveals signatures characterizing histologic subtypes of hepatoblastoma and global deregulation in cell growth and survival pathways. Hum Pathol 40(6):843–853
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM et al (2000) Gene ontology: tool for the unification of biology. Nat Genet 25(1):25–29
Bader GD, Hogue CW (2003) An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 4(1):2
Bandopadhyay M, Sarkar N, Datta S, Das D, Pal A, Panigrahi R et al (2016) Hepatitis B virus X protein mediated suppression of miRNA-122 expression enhances hepatoblastoma cell proliferation through cyclin G1-p53 axis. Infect Agents Cancer 11(1):40
Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M et al (2012) NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res 41(D1):D991–D995
Bell D, Ranganathan S, Tao J, Monga SP (2017) Novel advances in understanding of molecular pathogenesis of hepatoblastoma: a Wnt/β-catenin perspective. Gene Expression J Liver Res 17(2):141–154
Callegari E, Gramantieri L, Domenicali M, D'Abundo L, Sabbioni S, Negrini M (2015) MicroRNAs in liver cancer: a model for investigating pathogenesis and novel therapeutic approaches. Cell Death Differentiation 22(1):46–57
Cargnello M, Roux PP (2011) Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol Mol Biol Rev 75(1):50–83
Carrillo-Reixach J, Torrens L, Simon-Coma M, Royo L, Domingo-Sàbat M, Abril-Fornaguera J et al (2020) Epigenetic footprint enables molecular risk stratification of hepatoblastoma with clinical implications. J Hepatol 73:328–341
Cheah P-L, Looi L-M, Nazarina AR, Mun K-S, Goh K-L (2008) Association of Ki67 with raised transaminases in hepatocellular carcinoma. Malay 4:12.19
Chin C-H, Chen S-H, Wu H-H, Ho C-W, Ko M-T, Lin C-Y (2014) cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC systems biology 8(S4):S11
Chou C-H, Shrestha S, Yang C-D, Chang N-W, Lin Y-L, Liao K-W et al (2018) miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res 46(D1):D296–D302
Clarke DJB, Kuleshov MV, Schilder BM, Torre D, Duffy ME, Keenan AB et al (2018) eXpression2Kinases (X2K) web: linking expression signatures to upstream cell signaling networks. Nucleic Acids Res 46(W1):W171–W179
Czauderna P, Garnier H (2018) Hepatoblastoma: current understanding, recent advances, and controversies. F1000Research 7
Das T, Sangodkar J, Negre N, Narla G, Cagan R (2013) Sin3a acts through a multi-gene module to regulate invasion in Drosophila and human tumors. Oncogene 32(26):3184–3197
De Ioris M, Brugieres L, Zimmermann A, Keeling J, Brock P, Maibach R et al (2008) Hepatoblastoma with a low serum alpha-fetoprotein level at diagnosis: the SIOPEL group experience. Eur J Cancer 44(4):545–550
Dong R, Liu G, Liu B, Chen G, Li K, Zheng S, Dong K (2016) Targeting long non-coding RNA-TUG1 inhibits tumor growth and angiogenesis in hepatoblastoma. Cell Death Dis 7(6):e2278–e2278
Edgar R, Domrachev M, Lash AE (2002) Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30(1):207–210
Eichenmüller M, von Schweinitz D, Kappler R (2012) Therapeutic potential of CDK-inhibitors in hepatoblastoma. Klin Padiatr 224(03):A34
El-Rifai W (2014) AURKA regulates JAK2eSTAT3 activity in human gastric and esophageal cancers
Evan GI, Vousden KH (2001) Proliferation, cell cycle and apoptosis in cancer. Nature 411(6835):342–348
Feng J, Polychronidis G, Heger U, Frongia G, Mehrabi A, Hoffmann K (2019) Incidence trends and survival prediction of hepatoblastoma in children: a population-based study. Cancer Communications 39(1):62
Gao T, Han Y, Yu L, Ao S, Li Z, Ji J (2014) CCNA2 is a prognostic biomarker for ER+ breast cancer and tamoxifen resistance. PLoS One, 9(3)
Gao J, Li N, Dong Y, Li S, Xu L, Li X et al (2015) miR-34a-5p suppresses colorectal cancer metastasis and predicts recurrence in patients with stage II/III colorectal cancer. Oncogene 34(31):4142–4152
Goga A, Yang D, Tward AD, Morgan DO, Bishop JM (2007) Inhibition of CDK1 as a potential therapy for tumors over-expressing MYC. Nat Med 13(7):820–827
Goos JA, Coupé VM, Diosdado B, Delis-van Diemen PM, Karga C, Beliën JA et al (2013) Aurora kinase a (AURKA) expression in colorectal cancer liver metastasis is associated with poor prognosis. Br J Cancer 109(9):2445–2452
Griffiths EA, Pritchard S, McGrath S, Valentine HR, Price PM, Welch I, West CM (2008) Hypoxia-associated markers in gastric carcinogenesis and HIF-2α in gastric and gastro-oesophageal cancer prognosis. Br J Cancer 98(5):965–973
Halasi M, Gartel AL (2013) Targeting FOXM1 in cancer. Biochem Pharmacol 85(5):644–652
Han M-l, Wang F, Gu Y-t, Pei X-H, Ge X, Guo G-c et al (2016) MicroR-760 suppresses cancer stem cell subpopulation and breast cancer cell proliferation and metastasis: by down-regulating NANOG. Biomed Pharmacother 80:304–310
Hiyama E (2014) Pediatric hepatoblastoma: diagnosis and treatment. Transl Pediatr 3(4):293–299
Hu H, Li S, Liu J, Ni B (2012) MicroRNA-193b modulates proliferation, migration, and invasion of non-small cell lung cancer cells. Acta Biochim Biophys Sin 44(5):424–430
Kanehisa M, Goto S (2000) KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28(1):27–30
Koo C-Y, Muir KW, Lam EW-F (2012) FOXM1: from cancer initiation to progression and treatment. Biochimica et Biophysica Acta (BBA)-Gene Regul Mech 1819(1):28–37
Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, Monteiro CD, Gundersen GW, Ma'ayan A (2016) Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44(W1):W90–W97. https://doi.org/10.1093/nar/gkw377
Lee J, Choi C (2017) Oncopression: gene expression compendium for cancer with matched normal tissues. Bioinformatics 33(13):2068–2070
Loukil A, Cheung CT, Bendris N, Lemmers B, Peter M, Blanchard JM (2015) Cyclin A2: at the crossroads of cell cycle and cell invasion. World J Biol Chem 6(4):346–350
Magrelli A, Azzalin G, Salvatore M, Viganotti M, Tosto F, Colombo T et al (2009) Altered microRNA expression patterns in hepatoblastoma patients. Transl Oncol 2(3):157–163
Malumbres M, Pevarello P, Barbacid M, Bischoff JR (2008) CDK inhibitors in cancer therapy: what is next? Trends Pharmacol Sci 29(1):16–21
Manvati MKS, Khan J, Verma N, Dhar PK (2020) Association of miR-760 with cancer: an overview. Gene 144648
Mavila N, Thundimadathil J (2019) The emerging roles of Cancer stem cells and Wnt/Beta-catenin signaling in Hepatoblastoma. Cancers 11(10):1406
Mei Y, Yang J-P, Qian C-N (2017) For robust big data analyses: a collection of 150 important pro-metastatic genes. Chin J Cancer 36(1):16
Mokhlesi A, Talkhabi M (2020) Comprehensive transcriptomic analysis identifies novel regulators of lung adenocarcinoma. J Cell Commun Signal
Morgan DO (1997) Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu Rev Cell Dev Biol 13(1):261–291
Mrena J, Wiksten JP, Kokkola A, Nordling S, Haglund C, Ristimäki A (2006) Prognostic significance of cyclin a in gastric cancer. Int J Cancer 119(8):1897–1901
Nikonova AS, Astsaturov I, Serebriiskii IG, Dunbrack RL, Golemis EA (2013) Aurora a kinase (AURKA) in normal and pathological cell division. Cell Mol Life Sci 70(4):661–687
Otto T, Sicinski P (2017) Cell cycle proteins as promising targets in cancer therapy. Nat Rev Cancer 17(2):93–115
Pagano M, Pepperkok R, Verde F, Ansorge W, Draetta G (1992) Cyclin a is required at two points in the human cell cycle. EMBO J 11(3):961–971
Panvichian R, Tantiwetrueangdet A, Angkathunyakul N, Leelaudomlipi S (2015) TOP2A amplification and overexpression in hepatocellular carcinoma tissues. BioMed Res Int
Pearson G, Robinson F, Beers Gibson T, Xu B-e, Karandikar M, Berman K, Cobb MH (2001) Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr Rev 22(2):153–183
Saigusa S, Tanaka K, Mohri Y, Ohi M, Shimura T, Kitajima T et al (2015) Clinical significance of RacGAP1 expression at the invasive front of gastric cancer. Gastric Cancer 18(1):84–92
Semeraro M, Branchereau S, Maibach R, Zsiros J, Casanova M, Brock P et al (2013) Relapses in hepatoblastoma patients: clinical characteristics and outcome–experience of the international childhood liver tumour strategy group (SIOPEL). Eur J Cancer 49(4):915–922
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13(11):2498–2504
Sharp DJ, Rogers GC, Scholey JM (2000) Microtubule motors in mitosis. Nature 407(6800):41–47
Shin E, Lee K-B, Park S-Y, Kim S-H, Ryu H-S, Park Y-N, Yu E, Jang J-J (2011) Gene expression profiling of human hepatoblastoma using archived formalin-fixed and paraffin-embedded tissues. Virchows Arch 458(4):453–465
Smallridge RC, Marlow LA, Copland JA (2009) Anaplastic thyroid cancer: molecular pathogenesis and emerging therapies. Endocr Relat Cancer 16(1):17–44
Sumazin P, Chen Y, Treviño LR, Sarabia SF, Hampton OA, Patel K et al (2017) Genomic analysis of hepatoblastoma identifies distinct molecular and prognostic subgroups. Hepatology 65(1):104–121
Sun D, Lu J, Hu C, Zhang Q, Wang X, Zhang Z, Hu S (2018) Prognostic role of miR-760 in hepatocellular carcinoma. Oncol Lett 16(6):7239–7244
Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J et al (2019) STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47(D1):D607–D613
Talkhabi M, Razavi SM, Salari A (2017) Global transcriptomic analysis of induced cardiomyocytes predicts novel regulators for direct cardiac reprogramming. J Cell Commun Signal 11(2):193–204
van de Weerdt BC, Medema RH (2006) Polo-like kinases: a team in control of the division. Cell Cycle 5(8):853–864
Von Minckwitz G, Sinn H-P, Raab G, Loibl S, Blohmer J-U, Eidtmann H et al (2008) Clinical response after two cycles compared to HER2, Ki-67, p53, and bcl-2 in independently predicting a pathological complete response after preoperative chemotherapy in patients with operable carcinoma of the breast. Breast Cancer Res 10(2):R30
Wang SM, Ooi LLP, Hui KM (2007) Identification and validation of a novel gene signature associated with the recurrence of human hepatocellular carcinoma. Clin Cancer Res 13(21):6275–6283
Wang Z, Fan M, Candas D, Zhang T-Q, Qin L, Eldridge A et al (2014) Cyclin B1/Cdk1 coordinates mitochondrial respiration for cell-cycle G2/M progression. Dev Cell 29(2):217–232
Wong N, Yeo W, Wong WL, Wong NLY, Chan KYY, Mo FKF et al (2009) TOP2A overexpression in hepatocellular carcinoma correlates with early age onset, shorter patients survival and chemoresistance. Int J Cancer 124(3):644–652
Xu C, Liu S, Fu H, Li S, Tie Y, Zhu J, Xing R, Jin Y, Sun Z, Zheng X (2010) MicroRNA-193b regulates proliferation, migration and invasion in human hepatocellular carcinoma cells. Eur J Cancer 46(15):2828–2836
Yamada S-i, Ohira M, Horie H, Ando K, Takayasu H, Suzuki Y et al (2004) Expression profiling and differential screening between hepatoblastomas and the corresponding normal livers: identification of high expression of the PLK1 oncogene as a poor-prognostic indicator of hepatoblastomas. Oncogene 23(35):5901–5911
Yang Z, Deng Y, Zhang K, Bai Y, Zhu J, Zhang J, Xin Y, Li L, He J, Wang W (2020) LIN28B gene polymorphisms modify hepatoblastoma susceptibility in Chinese children. J Cancer 11(12):3512–3518
Zhan T, Rindtorff N, Boutros M (2017) Wnt signaling in cancer. Oncogene 36(11):1461–1473
Zhang HF, Wang YC, Han YD (2018a) MicroRNA-34a inhibits liver cancer cell growth by reprogramming glucose metabolism. Mol Med Rep 17(3):4483–4489
Zhang L, Jin Y, Zheng K, Wang H, Yang S, Lv C et al (2018b) Whole-genome sequencing identifies a novel variation of was gene coordinating with heterozygous Germline mutation of Apc to enhance Hepatoblastoma Oncogenesis. Front Genet 9:668
Zhang Y, Zhao Y, Wu J, Liangpunsakul S, Niu J, Wang L (2018c) MicroRNA-26-5p functions as a new inhibitor of hepatoblastoma by repressing lin-28 homolog B and aurora kinase a expression. Hepatol Commun 2(7):861–871
Acknowledgments
This study was supported by a grant from Shahid Beheshti University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests
The authors declare that there are no conflict of interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Aghajanzadeh, T., Tebbi, K. & Talkhabi, M. Identification of potential key genes and miRNAs involved in Hepatoblastoma pathogenesis and prognosis. J. Cell Commun. Signal. 15, 131–142 (2021). https://doi.org/10.1007/s12079-020-00584-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12079-020-00584-1