Abstract
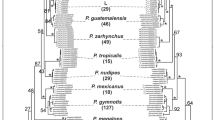
The species of harvest mouse Reithrodontomys megalotis belongs to one of the most diverse genera in North America with a wide latitudinal and altitudinal distribution. Previous studies suggest the existence of great morphological, chromosomal, and genetic variability; however, few of them have addressed the evolutionary patterns at the intraspecific level. We analyzed the evolutionary relationships and levels of genetic divergence in R. megalotis using DNA sequences of the mitochondrial cytochrome b gene. The results of the analysis with different phylogeographic methods show that R. megalotis represents a monophyletic group with respect to R. zacatecae, and populations included here split in three groups that were always consistent. Populations from the southern distribution form the first clade; from western North America form the second clade; and the third one comprises the populations of the Central Plains from the southern United States and the Mexican Plateau. The values of average genetic divergence within and among clades are low (< 3.4 %) compared to other species with similar distribution and times of divergence and origin. The results of this work are discussed in the context of the low genetic divergence found, given its wide and heterogeneous distribution together with its morphological variation. It is suggested that populations of R. megalotis have probably developed considerable phenotypic plasticity whose analysis and description should be approached from an ecological more than historical perspective.




Similar content being viewed by others
References
Akaike H (1973) Information theory and an extension of the maximum likelihood principle. In: Petrov BN, Caski F (eds) Proceedings of the Second International Symposium on Information Theory. Akadémiai Kiadó, Budapest, pp 267–281
Akaike H (1974) A new look at the statistical model identification. IEEE Trans Autom Contr 19:716–723
Albright LB (1999) Magnetostratigraphy and biochronology of the San Timoteo Badlands, southern California, with implications for local Pliocene-Pleistocene tectonic and depositional patterns. Geol Soc Am Bull 111:1265–1293
Alvarez-Castañeda ST, Rios E (2003) Noteworthy record of Western harvest mouse (Muridae: Reithrodontomys megalotis) on the Baja California Peninsula. Southwest Nat 48:471–472
Arbogast BS, Edwards SV, Wakeley J, Beerli P, Slowinski JB (2002) Estimating divergence times from molecular data on phylogenetic and population genetic timescales. Annu Rev Ecol Evol Syst 33:707–740
Arellano E, González-Cozátl F, Rogers DS (2005) Molecular systematics of Middle American harvest mice Reithrodontomys (Muridae), estimated from mitochondrial cytochrome b gene sequences. Mol Phylogenet Evol 37:529–540
Baird SF 1857 (1858) Mammals. In: Reports of Explorations and Surveys, to Ascertain the Most Practicable and Economical Route for a Railroad from the Mississippi River to the Pacific Ocean. Beverly Tucker Printer, Washington, D.C., 8(1):1–757 + 43 plates
Bandelt HJ, Forster P, Sykes BC, Richards MB (1995) Mitochondrial portraits of human populations. Genetics 141:743–753
Bradley RD, Baker RJ (2001) A test of the genetic species concept: Cytochrome b sequences and mammals. J Mammal 82:960–973
Bell DM, Hamilton MJ, Edwards CW, Wiggins LE, Muniz Martinez R, Strauss RE, Bradley RD, Baker RJ (2001) Patterns of karyotypic megaevolution in Reithrodontomys: evidence from a cytochrome-b phylogenetic hypothesis. J Mammal 82:81–91
Cassens I, Waerebeck KV, Best PB, Crespo EA, Reyes J, Milinkovitch MC (2003) The phylogeography of dusky dolphins (Lagenorhynchus obscurus): a critical examination of network methods and rooting procedures. Mol Ecol 12:1781–1792
Crandall KA (1996) Multiple interspecies transmissions of human and simian T-cell leukemia/lymphoma virus type I sequences. Mol Biol Evol 13: 115–131
Crandall KA, Templeton AR (1996) Applications of intraspecific phylogenetics. In: Harvey PH, Leigh Brown AJ, Maynard Smith J, Nee S (eds) New Uses for New Phylogenies. Oxford University Press, New York, pp 81–99
Dalquest WW (1975) Vertebrate fossils from the Blanco Local fauna of Texas. Occas Pap Mus Texas Tech Univ 30:1–52
Dalquest WW (1978) Early Blancan mammals of the Beck Ranch local fauna of Texas. J Mammal 59:269–298
Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9:772
Demboski JR, Sullivan J (2003) Extensive mtDNA variation within the yellow-pine chipmunk, Tamias amoenus (Rodentia: Sciuridae), and phylogeographic inferences for northwest North America. Mol Phylogenet Evol 26:389–408
Drummond AJ, Ho SY, Phillips MJ, Rambaut A (2006) Relaxed phylogenetics and dating with confidence. PLoS Biol 4(5):e88
Drummond AJ, Suchard MA, Xie D, Rambaut A (2012) Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29:1969–1973
Edwards CW, Bradley RD (2002) Molecular systematics and the historical phylobiogeography of the Neotoma mexicana species group. J Mammal 83:20–30
Erlich HA (1989) PCR technology. In: Erlich HA (ed) Principles and Applications for DNA Amplification, Stockton Press, New York, pp 246–252
Eshelman RE (1975) Geology and paleontology of the early Pleistocene (late Blancan) White Rock fauna from north-nentral Kansas. In: Studies on Cenozoic Paleontology and Stratigraphy, Claude W. Hibbard Memorial Vol., 4, Univ Mich Papers Paleontol 13:1–60
Excoffier L, Lischer HE (2010) Arlequin suite ver. 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Ford SD (1977) Range, distribution and habitat of the western harvest mouse, Reithrodontomys megalotis, in Indiana. Am Midland Nat 98:422–432
Fu YX (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147:915–925
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321
Hall ER (1981) The Mammals of North America, 2nd ed. John Wiley and Sons, Inc., New York
Hardy DK, González-Cozatl F, Arellano E, Rogers D (2013) Molecular phylogeographic structure and phylogenetics of Sumichrast’s harvest mouse (Reithrodontomys sumichrasti: Family Cricetidae) based on mitochondrial and nuclear DNA sequences. Mol Phylogenet Evol 68:282–292
Harpending HC, Batzer MA, Gurven M, Jorde LB, Rogers AR, Sherry ST (1998) Genetic traces of ancient demography. Proc Natl Acad Sci USA 95:1961–1967
Harris D, Rogers DS, Sullivan J (2000) Phylogeography of Peromyscus furvus (Rodentia: Muridae) based on cytochrome b sequence data. Mol Ecol 9:2129–2135
Hibbard CW, Taylor DW (1960) Two late Pleistocene faunas from southwestern Kansas. Contrib Mus Paleontol Univ Mich 16:1–223
Hillis DM, Bull JJ (1993) An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst Biol 42:182–192
Hood CG, Robbins LW, Baker RJ, Shellhammer HS (1984) Chromosomal studies and evolutionary relationships of an endangered species, Reithrodontomys raviventris. J Mammal 65:655–667
Hooper ET (1952) A systematic review of harvest mice (genus Reithrodontomys) of Latin America. Misc Publ Mus Zool Univ Mich 77:1–255
Hornsby A, Matocq M (2012) Differential regional response of the bushy-tailed woodrat (Neotoma cinerea) to late Quaternary climate change. J Biogeog 39:289–305
Irwin DM, Kocher TD, Wilson AC (1991) Evolution of the cytochrome b gene of mammals. J Mol Evol 32:128–144.
Kerhoulas NJ, Arbogast BS (2010) Molecular systematic and Pleistocene biogeography of Mesoamerican flying squirrels. J Mammal 91:654–667
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
León-Paniagua L, Navarro-Sigüenza AG, Hernández-Baños B, Morales JC (2007) Diversification of the arboreal mice of the genus Habromys (Rodentia: Cricetidae: Neotominae) in the Mesoamerican highlands. Mol Phylogenet Evol 42:653–664
Librado P, Rozas J (2009) DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Malaney JL (2013) Exploring signals of historical demography in boreal mammals through integration of statistical conservation phylogenetics, taxonomy, and comparative phylogeography. Dissertation: Doctor of Philosophy, Biology, The University of New Mexico, Albuquerque
Malaney JL, Conroy CJ, Moffitt LA, Spoonhunter HD, Patton JL, Cook JA (2013) Phylogeography of the western jumping mouse (Zapus princeps) detects deep and persistent allopatry with expansion. J Mammal 94(5):1016–1029
McKnight ML (2005) Phylogeny of the Perognathus longimembris species group based on mitochondrial cytochrome-b: how many species? J Mammal 86:826–832
Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Gateway Computing Environments Workshop (GCE). IEEE, pp 1–8
Neiswenter SA, Riddle BR (2010) Diversification of the Perognathus flavus species group in emerging arid grasslands of western North America. J Mammal 91:348–362
Peralta-Garcia A, Samaniego-Herrera A, Valdez-Villavicencio JH (2007) Western harvest mouse, Reithrodontomys megalotis (Rodentia: Muridae), on Magdalena Island, Mexico. Southwest Nat 52:595–623
Posada D, Buckley TR (2004) Model selection and model averaging in phylogenetics: advantages of Akaike Information Criterion and Bayesian Approaches over likelihood ratio tests. Syst Biol 53:793–808
Rambaut A, Drummond AJ (2009) Tracer v1.5. Available from http://beast.bio.ed.ac.uk/Tracer
Rambaut A, Suchard MA, Xie D, Drummond AJ (2014) Tracer v1.6. Available from http://beast.bio.ed.ac.uk/Tracer
Rogers AR, Harpending H (1992) Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol 9:552–569
Rogers DS, Arellano E, González-Cózatl FX, Hardy DK, Hanson JD, Lewis-Rogers N (2009) Molecular phylogenetics of Oligoryzomys fulvescens based on cytochrome b gene sequences, with comments on the evolution of the genus Oligoryzomys. In: Cervantes FA, Hortelano MY, Vargas CJ (eds) 60 años de la Colección Nacional de Mamíferos del Instituto de Biología, UNAM. Aportaciones al Conocimiento y Conservación de los Mamíferos Mexicanos. Universidad Autónoma de México, México, D. F, pp 209–222
Rogers DS, Funk CC, Miller JR, Engstrom MD (2007) Molecular phylogenetic relationships among crested-tailed mice (genus Habromys). J Mammal Evol 14:37–55
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61(3): 539–542
Ruff S, Wilson D (1999) The Smithsonian Book of North American Mammals. Smithsonian Institution Press in association with the American Society of Mammalogist, Washington, D.C.
Ruiz JC (2002) Filogeografía de una especie de roedor múrido endémico al Eje Neo volcánico Transversal (Reithrodontomys chrysopsis). Tesis de licenciatura. Universidad Nacional Autónoma de México. México.
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sánchez O (1993) Análisis de algunas tendencias ecogeográficas del género Reithrodontomys (Rodentia: Muridae) en México. In: Medellín RA, Cevallos G (eds) Avances en el estudio de los mamíferos de México, Publicaciones Especiales, Vol. 1, Asociación Mexicana de Mastozoología, A. C. México, D. F, pp 25–44
Schultz GE (1969) Geology and paleontology of a late Pleistocene basin in southwest Kansas. Geol Soc Am Spec Pap 105:1–85
Smith MF, Patton JL (1993) The diversification of South American murid rodents: evidence from mitochondrial DNA sequence data for the Akodontine tribe. Biol J Linn Soc 50:149–177
Sullivan J, Arellano E, Rogers DS (2000) Comparative phylogeography of Mesoamerican Highland rodents: concerted versus independent response to past climatic fluctuations. Am Nat 155:755–768
Sullivan J, Markert JA, Kilpatrick CW (1997) Phylogeography and molecular systematics of the Peromyscus aztecus species group (Rodentia: Muridae) inferred using parsimony and likelihood. Syst Biol 46:426–440
Swofford DL (2003) PAUP*. Phylogenetic analysis using parsimony (*and other methods) Version 4.0b10. Sinauer Associates, Inc., Sunderland, Massachusetts.
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595
Tomida Y (1987) Small mammal fossils and correlation of continental deposits, Safford and Duncan basins, Arizona, USA. National Science Museum, Tokyo. Science Museum Monographs 1–141
Urbina SI (2011) Variación cromosómica de especies del género Reithrodontomys de Bosque Mesófilo de Montaña. Tesis de Maestría. Universidad Autónoma Metropolitana-Iztapalapa. México, D.F
Vallejo RM, González-Cózatl FX (2012) Phylogenetic affinities and species limits within the genus Megadontomys (Rodentia:Cricetidae) based on mitochondrial sequence data. J Zool Syst Evol Res 50(1):67–75
Wagner HM, Riney BO, Deméré TA, Prothero DR (2001) Magnetic stratigraphy and land mammal biochronology of a non-marine facies of the Pliocene San Diego Formation, San Diego County, California. In: Prothero DR (ed) Magnetic Stratigraphy of the Pacific Coast Cenozoic: Pacific Section SEPM Spec Publ 91:359–368
Whitaker JO, Mumford RE (1972) Ecological studies on Reithrodontomys megalotis in Indiana. J Mammal 53:850–860
Wright SD (1949) The genetical structure of populations. Annals of Eugenics 15:323–354
Yang Z, Rannala B (1997) Bayesian phylogenetic inference using DNA sequences: a Markov chain Monte Carlo method. Mol Biol Evol 14:717–724
Yasuda SP, Vogel P, Tsuchiya K, Han SH, Lin LK, Suzuki H (2005) Phylogeographic patterning of mtDNA in the widely distributed harvest mouse (Micromys minutus) suggests dramatic cycles of range contraction and expansion during the mid- to late Pleistocene. Can J Zool 83:1411–1420
Acknowledgments
This study is part of the PhD Dissertation of ENG, supported by the Consejo Nacional de Ciencia y Tecnología scholarship number 163092. We thank Drs. James L. Patton of the Museum of Vertebrate Zoology, University of California; Duke S. Rogers of the Monte L. Bean Life Science Museum, Brigham Young University; Sergio Ticul Álvarez-Castañeda of the Centro de Investigaciones Biológicas del Noroeste; Celia López González of the Centro Interdisciplinario de Investigación para el Desarrollo Integral Regional Unidad Durango; Francisco X. González-Cózatl of the Centro de Investigación en Biodiversidad y Conservación, Universidad Autónoma del Estado de Morelos for providing tissue samples and/or DNA sequences. We also thank Dr. Rachel M. Vallejo, Cyndi Colin L. Roman, Mariana González Tellez, Armando Pérez Barrios, Sergio Albino Miranda, and Scarleth Sotelo Reyes for their help doing field work; and Dr. Ana L. Almendra for her help and comments in some analyses. The research conducted with partial resources granted by the Universidad Autónoma del Estado de Morelos through Fondo de Consolidación para Universidades Públicas Estatales y Apoyo Solidario 2010. Finally, we appreciate the comments made by anonymous reviewers, which were useful to the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
ESM 1
(DOC 102 kb)
Rights and permissions
About this article
Cite this article
Nava-García, E., Guerrero-Enríquez, J.A. & Arellano, E. Molecular Phylogeography of Harvest Mice (Reithrodontomys megalotis) Based on Cytochrome b DNA Sequences. J Mammal Evol 23, 297–307 (2016). https://doi.org/10.1007/s10914-015-9318-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10914-015-9318-5