Abstract
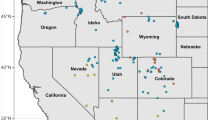
Chionactis occipitalis (Western Shovel-nosed Snake) is a small colubrid snake inhabiting the arid regions of the Mojave, Sonoran, and Colorado deserts. Morphological assessments of taxonomy currently recognize four subspecies. However, these taxonomic proposals were largely based on weak morphological differentiation and inadequate geographic sampling. Our goal was to explore evolutionary relationships and boundaries among subspecies of C. occipitalis, with particular focus on individuals within the known range of C. o. klauberi (Tucson Shovel-nosed snake). Population sizes and range for C. o. klauberi have declined over the last 25 years due to habitat alteration and loss prompting a petition to list this subspecies as endangered. We examined the phylogeography, population structure, and subspecific taxonomy of C. occipitalis across its geographic range with genetic analysis of 1100 bases of mitochondrial DNA sequence and reanalysis of 14 morphological characters from 1543 museum specimens. We estimated the species gene phylogeny from 81 snakes using Bayesian inference and explored possible factors influencing genetic variation using landscape genetic analyses. Phylogenetic and population genetic analyses reveal genetic isolation and independent evolutionary trajectories for two primary clades. Our data indicate that diversification between these clades has developed as a result of both historical vicariance and environmental isolating mechanisms. Thus these two clades likely comprise ‘evolutionary significant units’ (ESUs). Neither molecular nor morphological data are concordant with the traditional C. occipitalis subspecies taxonomy. Mitochondrial sequences suggest specimens recognized as C. o. klauberi are embedded in a larger geographic clade whose range has expanded from western Arizona populations, and these data are concordant with clinal longitudinal variation in morphology.





Similar content being viewed by others
References
Alfaro ME, Zoller S, Lutzoni F (2003) Bayes or bootstrap? A simulation study comparing the performance of Bayesian Markov chain Monte Carlo sampling and bootstrapping in assessing phylogenetic confidence. Mol Biol Evol 20:255–266
Anderson MJ (2004) DISTLM Version 5: a FORTRAN computer program to calculate a distance-based multivariate analysis for a linear model. Department of Statistics, University of Auckland, New Zealand. http://www.stat.auckland.ac.nz/~mja. Cited 16 Jun 2005
Avise JC (1994) Molecular markers, natural history and evolution. Chapman & Hall, New York
Avise JC (1996) Toward a regional conservation genetics perspective: phylogeography of faunas in the southeastern United States. In: Avise JC, Hamrick JL (eds) Conservation genetics: case histories from nature. Chapman and Hall, New York, pp 431–470
Ayoub NA, Riechert SE (2004) Molecular evidence for Pleistocene glacial cycles driving diversification of a North American desert spider, Agelenopsis aperta. Mol Ecol 13:3453–3465
Babik W, Branicki W, Crnobrnja-Isailovic J et al (2005) Phylogeography of the two European newt species-discordance between mtDNA and morphology. Mol Ecol 14: 2475–2491
Bailey RG, Avers PE, King T et al (eds) (1994) Ecoregions and subregions of the United States (map). Washington, DC, US Geological Survey. Scale 1:7,500,000. Accompanied by a supplementary table of map unit descriptions, McNab WH, Bailey RG (eds). Prepared for the USDA, Forest Service
Bethancourt JL (2004) Arid lands paleobiogeography: the rodent midden record in the Americas. In: Lomolino MV, Heaney LR (eds) Frontiers in biogeography. Sinauer Associates, Sunderland MA, pp 27–65
Brandley MC, Schmitz A, Reeder TW (2005) Partitioned Bayesian analyses, partition choice, and phylogenetic relationships of scincid lizards. Syst Biol 54:373–390
Center for Biological Diversity (2004) Petition to list the Tucson Shovel-nosed snake (Chionactis occipitalis klauberi) as an endangered species. Available from http://www.biologicaldiversity.org/swcbd. Cited 15 Dec 2004
Clement M, Posada D, Crandall KA (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9:1657–1659
Coachella valley multiple species habitat conservation plan (2006). Available from http://www.cvmshcp.org/prdplan/CVMSHCP_final.htm
Coyn JA, Orr HA (2004) Speciation. Sinauer Associates Inc., Boston
Crandall KA, Beninda-Emonds ORP, Mace GM et al (2000) Considering evolutionary processes in conservation biology. TREE 15:290–295
Cross JK (1979) Multivariate and univariate character geography in Chionactis (Reptilia: Serpentes). PhD Thesis, The University of Arizona
Dodd CK Jr, Seigel RA (1991) Relocation, repatriation, and translocation of amphibians and reptiles: Are they conservation strategies that work? Herpetologica 47:336–350
Douglas ME, Douglas MR, Schuett GW et al (2002) Phylogeography of the Western Rattlesnake (Crotalus viridis) complex, with emphasis on the Colorado Plateau. In: Shuett GW, Hoggren M, Douglas ME, Greene HW (eds) Biology of the Vipers. Eagle Mountain Publishing, LC, Eagle Mountain, UT, pp. 11–50
Douglas ME, Douglas MR, Schuett GW et al (2006) Evolution of rattlesnakes (Viperidae; Crotalus) in the warm deserts of western North America shaped by Neogene vicariance and Quaternary climate change. Mol Ecol 15:3353–3374
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol Bioinform Online 1:47–50
Felsentein J (1985) Confidence limits on phylogenies: an approach using bootstrap. Evolution 39:783–791
Fraser DJ, Bernatchez L (2001) Adaptive evolutionary conservation: towards a unified concept for identifying conservation units. Mol Ecol 10:2741–2752
Funk DJ, Omland KE (2003) Species-level paraphyly and polyphyly: frequency, causes, and consequences, with insights from animal mitochondrial DNA. Annu Rev Ecol Syst 34:397–423
Gatesy J, Desalle R, Wheeler W (1993) Alignment-ambiguous nucleotide sites and the exclusion of systematic data. Mol Phylogenet Evol 2:152–157
Geffen E, Anderson MJ, Wayne RK (2004) Climate and habitat barriers to dispersal in the highly mobile grey wolf. Mol Ecol 13:2481–2490
Goldstein PZ, Desalle R, Amato G et al (2000) Conservation genetics at the species boundary. Conserv Biol 14:120–131
Haig SM, Beever EA, Chambers SM et al (2006) Taxonomic considerations in listing subspecies under the U.S. Endangered Species Act Conserv Biol 20:1584–1594
Hillis DM, Bull JJ (1993) An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst Biol 42:182–192
Hillis DM, Mable BK, Larson A et al (1996) Nucleic acids IV: Sequencing and cloning, In: Hillis DM, Moritz C, Mable BK (eds) Molecular systematics, 2nd edn. Sinauer, Sunderland, Massachusetts, pp 231–381
Holycross AT, Douglas ME (2007) Geographic isolation, genetic divergence, and ecological non-exchangeability define ESUs in a threatened sky-island rattlesnake. Biol Conserv 134:142–154
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inferences of phylogenetic trees. Bioinform 17:754–755
Irwin DE (2002) Phylogeographic breaks without geographic barriers to gene flow. Evolution 56:2383–2394
Janzen FJ, Krenz JG, Haselkorn TS et al (2002) Molecular phylogeography of common garter snakes (Thamnophis sirtalis) in western North America: implications for regional historical forces. Mol Ecol 11:1739–1751
Jensen JL, Bohonak AJ, Kelley ST (2005) Isolation by distance, web service. BMC Genet 6:13
Kimura M (1980) A simple method of estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
King RB, Lawson R (1995) Color-pattern variation in Lake Erie water snakes: the role of gene flow. Evolution 49:885–896
Klauber LM (1937) A new snake of the genus Sonora from Mexico. Trans San Diego Soc Nat Hist 8:363–366
Klauber LM (1939) Studies of reptile life in the arid southwest. Bull Zoo Soc San Diego 14:65–79
Klauber LM (1951) The shovel-nosed snake, Chionactis with descriptions of two new subspecies. Trans San Diego Soc Nat Hist 11:141–204
Leaché AD, Reeder TW (2002) Molecular Systematics of the eastern fence lizard (Sceloporus undulatus): a comparison of parsimony, likelihood, and bayesian approaches. Sys Biol 51:44–68
Legendre P, Anderson MJ (1999) Distance based redundancy analysis: testing multispecies responses in multifactorial ecological experiments. Ecol Monogr 69:1–24
Mahrdt CR, Beaman KR, Rosen PC et al (2001) Chionactis occipitalis. Catalog Am Amphib Reptile 731:1–12
McArdle BH, Anderson MJ (2001) Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology 82:290–297
McCune B, Mefford MJ (1999) PC-ORD: Multivariate analysis of ecological data version 4.26. MjM Software Design, Gleneden Beach, Oregon
Miller MP (2005) Alleles in space (AIS): Computer software for the joint analysis of interindividual spatial and genetic information. J Hered 96:722–724
Miller MP, Bellinger R, Forsman ED, Haig SM (2006) Effects of historical climate change, habitat connectivity, and vicariance on genetic structure and diversity across the range of the red tree vole (Phenacomys longicaudus) in the Pacific Northwestern United States. Mol Ecol 15:145–159
Mortiz C (1994) Applications of mitochondrial DNA analysis in conservation: A critical review. Mol Ecol 3:401–411
Murphy RW, Trépanier TL, Morafka DA (2007) Conservation genetics, evolution and distinct population segments of the Mojave fringe-toed lizard, Uma scoparia. J Arid Environ 67:226–247
Nylander JAA (2002) MrModeltest v1.0b. Program distributed by the author via http://www.abc.se/~nylander/. Cited 27 Apr 2007
Nylander JAA, Ronquist F, Huelsenbeck JP et al (2004) Bayesian phylogenetic analysis of combined data. Syst Biol 53:47–67
Osada N, Wu C (2005) Inferring mode of speciation from genomic data: a study of the Great Apes. Genetics 169:259–264
Pilot M, Jedrezejewski W, Branicki W et al (2006) Ecological factors influence population genetic structure of European grey wolves. Mol Ecol 15:4533–4553
Posada D, Crandall KA, Templeton AR (2000) GEODIS: a program for the cladistic nested analysis of the geographical distribution of genetic haplotypes. Mol Ecol 9:487–488
Rosen PC (2003) Avra Valley snakes: Marana survey report for Tucson Shovel-nosed snake (Chionactis occipitalis klauberi). Final Report on shovel-nosed snake prepared for Town of Marana. University of Arizona, Tucson, AZ
Ryder OA (1986) Species conservation and the dilemma of subspecies. TREE 1:9–10
Schulte II JA, Macey JR, Papenfuss TJ (2006) A genetic perspective on the geographic association of taxa among arid North American lizards of the Sceloporus magister complex (Squamata: Iguanidae: Phrynosomatinae). Mol Phylogenet Evol 39:873–880
Shaw KL (2002) Conflict between nuclear and mitochondrial DNA phylogenies of a recent species radiation: what mtDNA reveals and conceals about modes of speciation in Hawaiian crickets. Proc Natl Acad Sci USA 99:16122–16127
Stickel WH (1941) The subspecies of the Spade-nosed snake, Sonora occipitalis. Bull Chicago Acad Sci 6:135–140
Swofford DL (2002) PAUP*: Phylogenetic Analysis Using Parsimony (* and Other Methods), Version 4.0b10. Sinauer, Sunderland, Massachusetts
Templeton AR (1998) Nested clade analyses of phylogeographic data: testing hypotheses about gene flow and population history. Mol Ecol 7:381–397
Templeton AR (2004) Statistical phylogeography: methods of evaluating and minimizing inference errors. Mol Ecol 13:789–809
Templeton AR, Crandall KA, Sing CF (1992) A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics 132:619–633
Thompson JD, Gibson TJ, Plewniak F et al (1997) The ClustalX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
Trépanier TL, Murphy RW (2001) The Coachella Valley Fringe-toed lizard Uma inornata: genetic diversity and phylogenetic relationships of an endangered species. Mol Phylogenet Evol 18:327–334
USFWS (U.S. Fish, Wildlife Service) and NMFS (National Marine Fisheries Service) (1996) Policy regarding the recognition of distinct vertebrate population segments under the Endangered Species Act. Fed Regist 61:4721–4725
Van Devender TM, Thompson RS, Betancourt JL (1987). Vegetation history of the deserts of southwestern North America; The nature and timing of the Late Wisconsin-Holocene transition. In: Ruddiman WF, Wright HE Jr (eds) North America and adjacent oceans during the last deglaciation. The Geology of North America K-3, Geological Society of America, Boulder, Colorado, pp 323–352
Wüster W, Yrausquin JL, Mijares-Urrutia (2001) A new species of indigo snake from north-western Venezuela (Serpentes: Colubridae: Drymarchon). Herpetol J 11:157–165
Acknowledgements
The Arizona Game and Fish Department (AGFD) provided funding that directly supported this project. Funding for tissue collections was received by USGS from Bureau of Land Management, Bureau of Recreation. We thank Bill Burger and Tom Jones for their field collection efforts and coordination of the volunteer collection efforts in Arizona. For contributing specimens and sharing information, data, and/or tissue samples we thank Randy Babb, Michael Barker, Kit Bezy, Robert Bezy, Valerie Boyarski, Thomas Brennan, Lisa Bucci, Kerry Crowther, Paul Daniel, Melanie Engstrom, Steve Goodman, Brad Hollingsworth, Larry Jones, Dominic Lanutti, Christine LiWanPo, Elizabeth Main, John Malone, Dan Mulcahy, Richard Myers, Lin Piest, Justin Pullins, Tanzy Pullins, Jim Rorabaugh, Phil Rosen, Angela Schwediman, Kirk Setser, Tim Snow, Donovan Wagner, Richard Wiggins, and the following institutions: San Diego Natural History Museum, Arizona State University. Collection was authorized by AGFD (SP613877, SP802036, SP572402), California Fish and Game Department (803038–04, 801135–04), and National Park Service (JOTR 25A0 9–07, JOTR-2005-SCI-0024). The use of trade, product, or firm names in this publication does not imply endorsement by the U.S. Government.
Author information
Authors and Affiliations
Corresponding author
Appendices
Rights and permissions
About this article
Cite this article
Wood, D.A., Meik, J.M., Holycross, A.T. et al. Molecular and phenotypic diversity in Chionactis occipitalis (Western Shovel-nosed Snake), with emphasis on the status of C. o. klauberi (Tucson Shovel-nosed Snake).. Conserv Genet 9, 1489–1507 (2008). https://doi.org/10.1007/s10592-007-9482-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-007-9482-0