Abstract
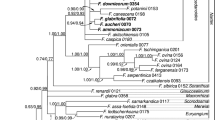
The present study, based on sequences of cpDNA (trnL-F & psbA-trnH) and nrDNA (ITS) and morphology, examined the evolutionary relationships in Blumea and its position among related genera. The results confirmed that the closest relatives of Blumea are Caesulia, Duhaldea and Pentanema p.p., and showed that the monotypic genera Blumeopsis and Merrittia are nested within Blumea. In Blumea s.l., two major, well-supported clades were recognised and a single species, the widespread Blumea balsamifera, that could not be placed with certainty relative to the two main clades. The two main clades differ in habit, ecology and distribution. The Blumea densiflora clade contains shrubs and subshrubs of evergreen forests, distributed from continental Asia to New Guinea and Polynesia, whereas the Blumea lacera clade is a widespread paleotropical group that comprises mostly annual, weedy herbs of open forests and fields.
Similar content being viewed by others
References
Álvarez I and Wendel JF (2003). Ribosomal ITS sequences and plant phylogenetic inference. Molec Phylogenet Evol 29: 417–434
Álvarez Fernández I, Fuertes Aguilar J, Panero JL and Nieto Feliner G (2001). A phylogenetic analysis of Doronicum (Asteraceae, Senecioneae) based on morphological, nuclear ribosomal (ITS), and chloroplast (trnL-F) evidence. Molec Phylogenet Evol 20: 41–64
Anderberg AA (1989). Phylogeny and reclassification of the tribe Inuleae (Asteraceae). Canad J Bot 67: 2277–2296
Anderberg AA (1991a). Taxonomy and phylogeny of the tribe Inuleae (Asteraceae). Pl Syst Evol 176: 75–123
Anderberg AA (1991b). Taxonomy and phylogeny of the tribe Plucheeae (Asteraceae). Pl Syst Evol 176: 145–177
Anderberg AA (1994). Tribe Inuleae. In: Bremer, K (eds) Asteraceae. Cladistics & classification, pp 273–291. Timber Press, Portland
Anderberg AA (1995). Doellia, an overlooked genus in the Asteraceae-Plucheeae. Willdenowia 25: 19–24
Anderberg AA and Eldenäs P (2007). XVII. Tribe Inuleae Cass. In: Kadereit, JW and Jeffrey, C (eds) The families and genera of vascular plants, vol 8. Flowering plants-eudicot, Asterales, pp 374–390. Springer, Berlin
Anderberg AA, Eldenäs P, Bayer R and Englund M (2005). Evolutionary relationships in the Asteraceae tribe Inuleae (incl. Plucheeae) evidenced by DNA sequences of ndhF; with notes on the systematic positions of some aberrant genera. Org Divers Evol 5: 135–146
Baldwin BG (1992). Phylogenetic utility of the internal transcribed spacers of nuclear ribosomal DNA in plants: an example from the Compositae. Molec Phylogenet Evol 1: 3–16
Bayer RJ and Cross EW (2003). A reassessment of tribal affinities of Cratystylis and Haegiela (Asteraceae) based on three chloroplast DNA sequences. Pl Syst Evol 236: 207–220
Bayer RJ and Starr JR (1998). Tribal phylogeny of the Asteraceae based on two non-coding chloroplast sequences, the trnL intron and trnL/trnF intergenic spacer. Ann Missouri Bot Gard 85: 242–256
Bentham G and Hooker JD (1873). Compositae. Plantarum. II. Genera Lovell Reeve, London
Bremer K (1994). Asteraceae. Cladistics & classification. Timber Press, Portland
Bremer B, Bremer K, Heidari N, Erixon P, Olmsted RG, Anderberg AA, Källersjö M and Barkhordarian E (2002). Phylogenetics of Asterids based on 3 coding and 3 non-coding chloroplast DNA marker and the utility of non-coding DNA at higher taxonomic levels. Molec Phylogenet Evol 24: 274–301
Burkill IH (1966). A dictionary of the economic products of the Malay Peninsula, vol I. The Ministry of Agriculture an co-operatives, Kuala Lumpur
de Candolle AP (1836). Prodr. V. Treuttel and Würtz, Paris
Clegg MT, Cummings MP and Durbin ML (1997). The evolution of plant nuclear genes. Proc Natl Acad Sci USA 94: 7791–7798
Dakshini KMM and Prithipalsingh (1977). Numerical taxonomy of the genus Blumea in India. Phytomorphism 27: 247–260
Dakshini KMM and Prithipalsingh (1978). Section Hieraciifoliae Randeria of Blumea DC. (Asteraceae-Inuleae). Taxon 27: 45–49
Doyle JJ (1992). Gene trees and species trees: molecular systematics as one-character taxonomy. Syst Bot 17: 144–163
Eldenäs PK, Anderberg AA and Källersjö M (1998). Molecular phylogenetics of the tribe Inuleae s.str. (Asteraceae), based on ITS sequences of nuclear ribosomal DNA. Pl Syst Evol 210: 159–173
Farris JS, Källersjö M, Kluge AG and Bult C (1994). Testing significance of incongruence. Cladistics 10: 315–319
Fehrer J, Gemeinholzer B, Chrtek J and Bräutigam S (2007). Incongruent plastid and nuclear DNA phylogenies reveal ancient intergeneric hybridization in Pilosella hawkweeds (Hieracium, Cichorieae, Asteraceae). Molec Phylogenet Evol 42: 347–361
Felsenstein J (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791
Francisco-Ortega J, Park S-J, Santos-Guerra A, Benabid A and Jansen RK (2001). Origin and evolution of the endemic Macaronesian Inuleae (Asteraceae): evidence from the internal transcribed spacers of nuclear ribosomal DNA. Biol J Linn Soc 72: 77–97
Gagnepain F (1920). Un genre nouveau de Composées. Bull Mus Nat Hist Par 26: 75–76
Goertzen LR, Cannone JJ, Gutell RR and Jansen RK (2003). ITS secondary structure derived from comparative analysis: implications for sequence alignment and phylogeny of the Asteraceae. Molec Phylogenet Evol 29: 216–234
Grierson AJC (1974). Critical notes on the Compositae of Ceylon, II. Ceylon J Sci 11: 12–15
Gustafsson MHG, Pepper AS-R, Albert VA and Källersjö M (2001). Molecular phylogeny of the Barnadesioideae (Asteraceae). Nord J Bot 21: 149–160
Hall TA (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41: 95–98
Karis PO (1993). Morphological phylogenetics of the Asteraceae-Asteroideae, with notes on character evolution. Pl Syst Evol 186: 69–93
Kim K-J and Jansen RK (1995). ndhF sequence evolution and the major clades in the sunflower family. Proc Natl Acad Sci USA 92: 10379–10383
Kim S-C, Crawford DJ, Jansen RK and Santos-Guerra A (1999). The use of non-coding region of chloroplast DNA in phylogenetic studies of the subtribe Sonchinae (Asteraceae: Lactuceae). Pl Syst Evol 215: 85–99
Kim S-C, Lu C-T and Lepschi BJ (2004). Phylogenetic positions of Actites megalocarpa and Sonchus hydrophilus (Sonchinae: Asteraceae) based on ITS and chloroplast non-coding DNA sequences. Austral Syst Bot 17: 73–81
Maddison WP and Maddison DR (2001). MacClade, analysis of phylogeny and character evolution, Version 4.03. Sinauer Associates, Sunderland
Mattfeld J (1929). Die Compositen von Papuasien. Bot Jahrb Syst 62: 425
McKenzie RJ, Muller EM, Skinner AKW, Karis PO and Barker NP (2006). Phylogenetic relationships and generic delimitation in subtribe Arctotidinae (Asteraceae: Arctotideae) inferred by DNA sequence data from ITS and five chloroplast regions. Amer J Bot 93: 1222–1235
Merrill ED and Merritt ML (1910). The flora of Mount Pulog. Philipp. J Sci 5: 393–401
Merxmüller H, Leins P and Roessler H (1977). Inuleae – systematic review. In: Heywood, VH, Harborne, JB, and Turner, BL (eds) The biology and chemistry of the Compositae II, pp 577–602. Academic, London
Noyes RD and Rieseberg LH (1999). ITS sequence data support a single origin for North American Astereae (Asteraceae) and reflect deep geographic divisions in Aster s.l. Amer. J Bot 86: 398–412
Pandey AK, Jha SM and Dhakal MR (2000). Seed coat, pericarp and pseudopericarp in Caesulia axillaris Roxb. (Asteraceae). Compositae Newslett 35: 37–43
Panero JL and Funk VK (2002). Toward a phylogenetic subfamilial classification for the Compositae (Asteraceae). Proc Biol Soc Washington 115: 909–922
Pelser PB, van den Hof K, Gravendeel B and van der Meijden R (2004). The systematic value of morphological characters in Senecio sect. Jacobaea (Asteraceae) as compared to DNA sequences. Syst Bot 29: 790–805
Randeria AJ (1960). The Compositae genus Blumea: A taxonomic revision. Blumea 10: 176–317
Sang T, Crawford DJ and Stuessy TF (1997). Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). Amer J Bot 84: 1120–1136
Schultz-Bipontinus CH (1843). Walpers, GG (eds) Reportorium Botanices Systematicae. Tomus II, pp. Sumtibus Friderici Hofmeister, Lipsiae
Simmons MP and Ochoterena H (2001). Gaps as characters in sequence-based phylogenetic analyses. Syst Biol 49: 369–381
Small RL, Cronn RC and Wendel JF (2004). L. A. S. Johnson review No. 2: Use of nuclear genes for phylogeny reconstruction in plants. Austral Syst Bot 17: 145–170
Soejima A and Wen J (2006). Phylogenetic analysis of the grape family (Vitaceae) based on three chloroplast markers. Amer J Bot 93: 278–287
Soltis DE, Johnson LA and Looney C (1996). Discordance between ITS and chloroplast topologies in the Boykinia Group (Saxifragaceae). Syst Bot 21: 169–185
Swofford DL (1998). PAUP*: Phylogenetic Analysis Using Parsimony (*and other methods), Version 4.0. Sinauer Associates, Sunderland
Taberlet P, Gielly L and Bouvet J (1991). Universal primers for amplification of three non-coding regions of chloroplast DNA. Pl Molec Biol 17: 1105–1109
Thompson JD, Higgins DG and Gibson TJ (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucl Acids Res 22: 4673–4680
Qiu Y-L, Lee J, Bernasconi-Quadroni F, Soltis DE, Soltis PS, Zanis M, Zimmer EA, Chen Z, Savolainenk V and Chase MW (1999). The earliest angiosperms: evidence from mitochondrial, plastid and nuclear genomes. Nature 402: 404–407
Wagstaff SJ and Breitwieser I (2002). Phylogenetic relationships of New Zealand Asteraceae inferred from ITS sequences. Pl Syst Evol 231: 203–224
Wild H (1969). The Compositae of the Flora Zambesiaca area II. Kirkia 7: 121–132
Wortley AH and Scotland RW (2006). The effect of combining molecular and morphological data in published phylogenetic analyses. Syst Biol 55: 677–685
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Pornpongrungrueng, P., Borchsenius, F., Englund, M. et al. Phylogenetic relationships in Blumea (Asteraceae: Inuleae) as evidenced by molecular and morphological data. Plant Syst. Evol. 269, 223–243 (2007). https://doi.org/10.1007/s00606-007-0581-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-007-0581-7