Abstract
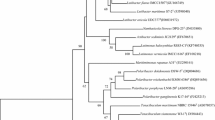
Strain S33T was isolated from oil-contaminated sediment of Tae-an coastal region of South Korea. Cells are aerobic, motile, Gram staining-negative, and coccoid shaped. Strain S33T grew optimally at the temperature of 25 °C (range of 4–40 °C), pH 6.0 (range of pH 6.0–10.0), and in the presence of 1 % (w/v) NaCl (range of 0–10 %). Ubiquinone-8 was the predominant respiratory quinone. C16:0, summed feature 3 (comprising C16:1ω7c/C16:1ω6c) and C18:1ω7c were the major fatty acids. The major polar lipids were phosphatidylethanolamine, phosphatidylglycerol, and diphosphatidylglycerol. Strain S33T showed the ability to degrade benzene, toluene, and ethylbenzene after 3 days incubation. 16S rRNA gene sequence analysis showed that the strain S33T was most closely related to Oceanisphaera sediminis TW92T (97.3 %), Oceanisphaera profunda SM1222T (97.2 %), and Oceanisphaera ostreae T-w6T (97.1 %) and <97 % with other members of the genus Oceanisphaera. The genomic DNA G+C mol% content of strain S33T was 51.0 mol%. Based on distinct phenotypic, genotypic, and phylogenetic analysis, strain S33T was proposed to represent a novel species in the genus Oceanisphaera as Oceanisphaera aquimarina sp. nov. (= KEMB 1002-058T = JCM 30 794T).

Similar content being viewed by others
Change history
27 November 2018
The original version of this article unfortunately contained a mistake in the provided KEMB accession number.
References
Cappuccino JG, Sherman N (2010) Microbiology: a laboratory manual, vol 69–74, 9th edn. Benjamin Cummings, San Francisco, pp 161–164
Choi WC, Kang SJ, Jung YT, Oh TK, Yoon JH (2011) Oceanisphaera ostreae sp. nov., isolated from seawater of an oyster farm, and emended description of the genus Oceanisphaera Romanenko et al. 2003. Int J Syst Evol Microbiol 61:2880–2884
Felsenstein J (1985) Confidence limit on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool 20:406–416
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41:95–98
Hiraishi A, Ueda Y, Ishihara J, Mori T (1996) Comparative lipoquinone analysis of influent sewage and activated sludge by high performance liquid chromatography and photodiode array detection. J Gen Appl Microbiol 42:457–469
Kimura M (1983) The neutral theory of molecular evolution. Cambridge University Press, Cambridge
Mesbah M, Premachandran U, Whitman WB (1989) Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int J Syst Bacteriol 39:159–167
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Met 2:233–241
Minnikin DE, Patel PV, Alshamaony L, Goodfellow M (1977) Polar lipid composition in the classification of Nocardia and related bacteria. Int J Syst Bacteriol 27:104–117
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucl Acids Res 8:4321–4326
Park SJ, Kang CH, Nam YD, Bae JW, Park YH, Quan ZX, Moon DS, Kim HJ, Roh DH, Rhee SK (2006) Oceanisphaera donghaensis sp. nov., a halophilic bacterium from the East Sea. Korea Int J Syst Evol Microbiol 56:895–898
Romanenko LA, Schumann P, Zhukova NV, Rohde M, Mikhailov VV, Stackebrandt E (2003) Oceanisphaera litoralis gen. nov., sp. nov., a novel halophilic bacterium from marine bottom sediments. Int J Syst Evol Microbiol 53:1885–1888
Satou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Shin NR, Whon TW, Roh SW, Kim MS, Kim YO, Bae JW (2012) Oceanisphaera sediminis sp. nov., isolated from marine sediment. Int J Syst Evol Microbiol 62:1926–1931
Srinivas TN, Reddy PV, Begum Z, Manasa P, Shivaji S (2012) Oceanisphaera arctica sp. nov., isolated from Arctic marine sediment, and emended description of the genus Oceanisphaera. Int J Syst Evol Microbiol 62:1926–1931
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Inc., Newark
Subhash Y, Sasikala Ch, Ramana CV (2013) Flavobacterium aquaticum sp. nov., isolated from a water sample of a rice field. Int J Syst Evol Microbiol 63:3463–3469
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
Tittsler RP, Sandholzer LA (1936) The use of semi-solid agar for the detection of bacterial motility. J Bacteriol 31:575–580
Xu Z, Zhang XY, Su HN, Yu ZC, Liu C, Li H, Chen XL, Song XY, Xie BB, Qin QL, Zhou BC, Shi M, Huang Y, Zhang YZ (2014) Oceanisphaera profunda sp. nov., a marine bacterium isolated from deep-sea sediment, and emended description of the genus Oceanisphaera. Int J Syst Evol Microbiol 64:1252–1256
Zhou S, Wang H, Wang Y, Ma K, He M, Chen X, Kong D, Guo X, Ruan Z, Zhao B (2015) Oceanisphaera psychrotolerans sp. nov., isolated from coastal sediment samples. Int J Syst Evol Microbiol 65:2797–2802
Acknowledgments
This research is supported by the National Research Foundation of Korea (NRF-2013M3A2A1067498) and (NRF-2015M3A9B8029697).
Author information
Authors and Affiliations
Corresponding author
Additional information
The GenBank/EMBL/DDBJ accession number for the 16S rRNA sequence of strain S33T (= KEMB 1002-058T = JCM 30794T) is KR003110.
Electronic Supplementary Material
Below is the link to the electronic supplementary material. Supplementary Fig. 1. Major polar lipids profiles using two-dimensional thin layer chromatography (TLC) of strain S33T (left) and O. sediminis TW92T. Diphosphatidylglycerol, phosphatidylglycerol, and phosphatidylethanolamine.
Rights and permissions
About this article
Cite this article
Cho, S., Lee, SS. Oceanisphaera aquimarina sp. nov., Isolated from Oil-Contaminated Sediment of Ocean Coastal Area from South Korea. Curr Microbiol 73, 618–623 (2016). https://doi.org/10.1007/s00284-016-1103-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-016-1103-z