Abstract
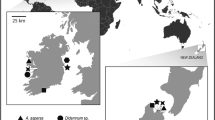
Bugula neritina is a common invasive cosmopolitan bryozoan that harbors (like many sessile marine invertebrates) a symbiotic bacterial (SB) community. Among the SB of B. neritina, “Candidatus Endobugula sertula” continues to receive the greatest attention, because it is the source of bryostatins. The bryostatins are potent bioactive polyketides, which have been investigated for their therapeutic potential to treat various cancers, Alzheimer’s disease, and AIDS. In this study, we compare the metagenomics sequences for the 16S ribosomal RNA gene of the SB communities from different geographic and life cycle samples of Chinese B. neritina. Using a variety of approaches for estimating alpha/beta diversity and taxonomic abundance, we find that the SB communities vary geographically with invertebrate and fish mariculture and with latitude and environmental temperature. During the B. neritina life cycle, we find that the diversity and taxonomic abundances of the SB communities change with the onset of host metamorphosis, filter feeding, colony formation, reproduction, and increased bryostatin production. “Ca. Endobugula sertula” is confirmed as the symbiont of the Chinese “Ca. Endobugula”/B. neritina symbiosis. Our study extends our knowledge about B. neritina symbiosis from the New to the Old World and offers new insights into the environmental and life cycle factors that can influence its SB communities, “Ca. Endobugula,” and bryostatins more globally.



Similar content being viewed by others
References
Ryland JS (1970) Bryozoans. Hutchinson, London
Hill K (2001) Bugula neritina. Smithsonian Marine Station at Fort Pierce. http://www.sms.si.edu/IRLSpec/Bugula_neriti.htm. Accessed 1 Mar 2018
Bishop Museum and University of Hawaii (2002) Guidebook of introduced marine species of Hawaii. Bugula neritina (Linnaeus, 1758). Hawaii Biological Survey, Honolulu
Haygood MG, Davidson SK (1997) Small-subunit rRNA genes and in situ hybridization with oligonucleotides specific for the bacterial symbionts in the larvae of the bryozoan Bugula neritina and proposal of “Candidatus Endobugula sertula”. Appl Environ Microbiol 63:4612–4616
Sharp KY, Davidson SK, Haygood MG (2007) Localization of ‘Candidatus Endobugula sertula’ and the bryostatins throughout the life cycle of the bryozoan Bugula neritina. ISME J 1:693–702
Lopanik NB (2015) Interactions between the marine bryozoan Bugula neritina, its endosymbiont, and symbiont-produced bryostatins. Planta Med 81:IL41
Davidson SK, Allen SW, Lim GE, Anderson CM, Haygood MG (2001) Evidence for the biosynthesis of bryostatins by the bacterial symbiont “Candidatus Endobugula sertula” of the bryozoan Bugula neritina. Appl Environ Microbiol 67:4531–4537
Yu X, Yan Y, Gu J-D (2007) Attachment of the biofouling bryozoan Bugula neritina larvae affected by inorganic and organic chemical cues. Int Biodeterior Biodegrad 60:81–89
Lopanik NB (2014) Chemical defense symbiosis in the marine environment. Funct Ecol 28:328–340
Mathew M, Lopanik NB (2014) Host differentially expressed genes during association with its defensive endosymbiont. Biol Bull 226:152–163
Gatley-Montross CM, Finlay JA, Aldred N, Cassady H, Destino JF, Orihuela B, Hickner MA, Clare AS, Rittschof D, Holm ER, Detty MR (2017) Multivariate analysis of attachment of biofouling organisms in response to material surface characteristics. Biointerphases 12:0511003
Lopanik N, Lindquist N, Targett N (2004) Potent cytotoxins produced by a microbial symbiont protect host larvae from predation. Oecologia 139:131–139
Lopanik NB, Targett NM, Lindquist N (2006) Ontogeny of a symbiont-produced chemical defense in Bugula neritina (Bryozoa). Mar Ecol Prog Ser 327:183–191
Trindade-Silva AE, Lim-Fong GE, Sharp KH, Haygood MG (2010) Bryostatins: biological context and biotechnological prospects. Curr Opin Biotechnol 21:834–842
Davidson SK, Haygood MG (1999) Identification of sibling species of the bryozoan Bugula neritina that produce different anticancer bryostatins and harbor distinct strains of the bacterial symbiont “Candidatus Endobugula sertula”. Biol Bull 196:273–280
McGovern TM, Hellberg ME (2003) Cryptic species, cryptic endosymbionts, and geographical variation in chemical defences in the bryozoan Bugula neritina. Mol Ecol 12:1207–1215
Mackie JA, Keough MJ, Christdis L (2006) Invasion patterns inferred from cytochrome oxidase I sequences in three bryozoans, Bugula neritina, Watersipora subtorquata, and Watersipora arcuata. Mar Biol 149:285–295
Linneman J, Paulus D, Lim-Fong G, Lopanik NB (2014) Latitudinal variation of a defensive symbiosis in the Bugula neritina (Bryozoa) sibling species complex. PLoS One 9:e108783
Pettit GR (1991) The bryostatins. Prog Chem Org Nat Prod 57:153–195
Lei H, Zhou X, Yang Y, Xu T, Yang X, Sun J, Yang B, Hu J, Lin X, Long L, Liu Y (2010) Bryostatins from South China Sea bryozoan Bugula neritina L. Biochem Syst Ecol 38:1231–1233
Yarza P, Yilmaz P, Pruesse E, Glöckner FO, Ludwig W, Schleifer K-H, Whitman WB, Euzéby J, Amann R, Rosselló-Móra R (2014) Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol 12:635–645
Caporaso JG, Christian LL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R (2011) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA 108:4516–4522
Miller IJ, Weyna TR, Fong SS, Lim-Fong GE, Kwan JC (2016) Single sample resolution of rare microbial dark matter in a marine invertebrate metagenome. Nat Sci Rep 6:34362
Huang ZG, Zheng CX, Lin S, Li CY, Wang JJ, Yan SK (1993) Fouling organisms at Daya Bay nuclear power station, China. In: Morton B (ed) The marine biology of the South China Sea: Proceedings of the First International Conference on the Marine Biology of Hong Kong and the South China Sea, Hong Kong, 28 October-3 November 1990, vol 1. Hong Kong University Press, Hong Kong, pp 121–130
Arnold AE, Miadlikowska J, Higgins KL, Sarvate SD, Gugger P, Way A, Hofstetter V, Kauff F, Lutzoni F (2009) A phylogenetic estimation of trophic transition networks for ascomycetous fungi: are lichens cradles of symbiotrophic fungal diversification? Syst Biol 58:283–297
Dahms H-U, Gao Q-F, Hwang J-S (2007) Optimized maintenance and larval production of the bryozoan Bugula neritina (Bugulidae: Gymnolaemata) in the laboratory. Aquaculture 265:169–175
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R (2012) Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624
Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD (2013) Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, Brown CT, Porras-Alfaro A, Kuskes CR, Tiedje JM (2014) Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res 42 (Database issue):D633–D642
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41 (Database issue):D590–D596
Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG (2013) Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods 10:57–59
Hartl DL, Clark AG (2007) Principles of population genetics, 4th edn. Sinauer, Sunderland
Hill TCJ, Walsh KA, Harris JA, Moffett BF (2003) Using ecological diversity measures with bacterial communities. FEMS Microbiol Ecol 43:1–11
Wong RG, Wu JR, Gloor GB (2016) Expanding the UniFrac toolbox. PLoS One 11:e0161196
Shannon CE (1948) A mathematical theory of communication. Bell Syst Tech J 27:379–423
Krebs CJ (1999) Ecological methodology, 2nd edn. Addison-Wesley, Menlo Park
Lozupone CA, Hamady M, Kelley ST, Knight R (2007) Quantitative and qualitative ß diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol 73:1576–1585
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267
Resource Coordinators NCBI (2016) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 44 (Database issue):D7–D19
Kim M, Oh H-S, Park S-C, Chun J (2014) Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol 64:346–351
Weiss S, Xu ZZ, Peddada S, Amir A, Bittinger K, Gonzalez A, Lozupone C, Zaneveld JR, Vázquez-Baeza Y, Birmingham A, Hyde ER, Knight R (2017) Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome 5:27
Schloss PD, Handelsman J (2005) Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol 71:1501–1505
Miller IJ, Vanee N, Fong SS, Lim-Fong GE, Kwan JC (2016) Lack of overt genome reduction in the bryostatin-producing bryozoan symbiont “Candidatus Endobugula sertula”. Appl Environ Microbiol 82:6573–6583
Lim GE, Haygood MG (2004) “Candidatus Endobugula glebosa,” a specific bacterial symbiont of the marine bryozoan Bugula simplex. Appl Environ Microbiol 70:4921–4929
Lim-Fong GE, Regali LA, Haygood MG (2008) Evolutionary relationships of “Candidatus Endobugula” bacterial symbionts and their Bugula bryozoan hosts. Appl Environ Microbiol 74:3605–3609
Zeng Y, Ma Y, Wei C, Jiao N, Tang K, Wu Z, Jian J (2010) Bacterial diversity in various coastal mariculture ponds in Southeast China and in diseased eels as revealed by culture and culture-independent molecular techniques. Aquac Res 41:e172–e186
Zhao W, Shen H (2016) A statistical analysis of China’s fisheries in the 12th five-year period. Aquacult Fisher 1:41–49
Cao L, Wang W, Yang Y, Yang C, Yuan Z, Xiong S, Diana J (2007) Environmental impact of aquaculture and countermeasures to aquaculture pollution in China. Environ Sci Pollut Res 14:452–462
Zhang Y, Bleeker A, Liu J (2015) Nutrient discharge from China’s aquaculture industry and associated environmental impacts. Environ Res Lett 10:045002
Wartenberg R, Feng L, Wu JJ, Mak YL, Chan LL, Telfer TC, Lam PKS (2017) The impacts of suspended mariculture on coastal zones in China and the scope for integrated multi-trophic aquaculture. Ecosyst Health Sustain 3:1340268
Halpern BS, Walbridge S, Selkoe KA, Kappel CV, Micheli F, D’Agrosa C, Bruno JF, Casey KS, Ebert C, Fox HE, Fujita R, Heinemann D, Lenihan HS, Madin EMP, Perry MT, Selig ER, Spalding M, Steneck R, Watson R (2008) A global map of human impact on marine ecosystems. Science 319:948–952
Ladau J, Sharpton TJ, Finucane MM, Jospin G, Kembel SW, O’Dwyer J, Koeppel AF, Green JL, Pollard KS (2013) Global marine bacterial diversity peaks at high latitudes in winter. ISME J 7:1669–1677
World sea temperature (2018) China sea temperatures. https://www.seatemperature.org/asia/china/. Accessed 1 Mar 2018
Fuhrman JA, Steele JA, Hewson I, Schwalbach MS, Brown MV, Green JL, Brown JH (2008) A latitudinal diversity gradient in planktonic marine bacteria. Proc Natl Acad Sci USA 105:7774–7778
Zhang M, Sun Y, Chen K, Yu N, Zhou Z, Chen L, Du Z, Li E (2014) Characterization of the intestinal microbiota in Pacific white shrimp, Litopenaeus vannamei, fed diets with different lipid sources. Aquaculture 434:449–455
Zhang M, Sun Y, Chen L, Cai C, Qiao F, Du Z, Li E (2016) Symbiotic bacteria in gills and guts of Chinese mitten crab (Eriocheir sinensis) differ from the free-living bacteria in water. PLoS One 11:e0148135
Carter RWG (1988) Coastal environments. An introduction to the physical, ecological and cultural systems of coastlines. Academic, London
Evans D (ed) (2009) Osmotic and ionic regulation: cells and animals, 1st edn. CRC Press, Boca Raton
Swezey DS, Bean JR, Hill TM, Gaylord B, Ninokawa AT, Sanford E (2017) Plastic responses of bryozoans to ocean acidification. J Exp Biol 220:4399–4409
Woolacott RM, Zimmer RL (1971) Attachment and metamorphosis of the cheilo-ctenostome bryozoan bugula neritina (linné). J Morphol 134:351–382
Wong YH, Ryu T, Seridi L, Ghosheh Y, Bougouffa S, Qian P-Y, Ravasi T (2014) Transcriptome analysis elucidates key developmental components of bryozoan lophophore development. Nat Sci Rep 4:6534
Barneche DR, White CR, Marshall DJ (2017) Temperature effects on mass-scaling exponents in colonial animals: a manipulative test. Ecology 98:103–111
Ostrovsky AN (2013) Evolution of sexual reproduction in marine invertebrates: example of gymnolaemate bryozoans. Springer, Dordrecht
Fehlauer-Ale KH, Mackie JA, Lim-Fong GE, Ale E, Pie MR, Waeschenbach A (2014) Cryptic species in the cosmopolitan Bugula neritina complex (Bryozoa, Cheilostomata). Zool Scr 43:193–205
Wong CS, Harrison PJ (eds) (1992) Marine ecosystem enclosed experiments. Proceedings of a symposium held in Beijing, People’s Republic of China, 9–14 May 1987. International Development Research Centre, Ottawa
Acknowledgements
We thank Charles Baer, Edward Braun, Keith P. Choe, Michele R. Tennant, and George P. Tiley for their helpful comments about this research, and Lin Xiangzhi and Niu Sufang for their aid in photography. This work was supported by funds from the Public Welfare Project of the State Oceanic Administration (Grant Number 201205024-2), National Basic Research Foundation of China (2013FY110700), and Department of Biology, University of Florida.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Li, H., Mishra, M., Ding, S. et al. Diversity and Dynamics of “Candidatus Endobugula” and Other Symbiotic Bacteria in Chinese Populations of the Bryozoan, Bugula neritina. Microb Ecol 77, 243–256 (2019). https://doi.org/10.1007/s00248-018-1233-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-018-1233-x