Abstract
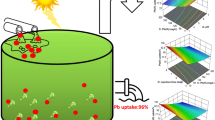
In this study, initial minimum inhibitory concentration (MIC) of Pb(II) ions was analysed to check optimum concentration of Pb(II) ions at which the growth of sulphate-reducing consortium (SRC) was found to be maximum. 80 ppm of Pb(II) ions was investigated as minimum inhibitory concentration for SRC. Influence of electron donors such as lactose, sucrose, glucose and sodium lactate was examined to investigate best carbon source for growth and activity of sulphate-reducing bacteria. Sodium lactate was found to be the prime carbon source for SRC. Later optimization of various parameters was executed using Box–Behnken design model of response surface methodology to explore the effectiveness of three independent operating variables, namely, pH (5.0–9.0), temperature (32–42 °C) and time (5.0–9.0 days), on dependent variables, i.e. protein content, precipitation of Pb(II) ions, and removal of COD by SRC biomass. Maximum removal of COD and Pb(II) was observed to be 91 and 98 %, respectively, at pH 7.0 and temperature 37 °C and incubation time 7 days. According to response surface analysis and analysis of variance, the experimental data were perfectly fitted to the quadratic model, and the interactive influence of pH, temperature and time on Pb(II) and COD removal was highly significant. A high regression coefficient between the variables and response (r 2 = 0.9974) corroborate eminent evaluation of experimental data by second-order polynomial regression model. SEM and Fourier transform infrared analysis was performed to investigate morphology of PbS precipitates, sorption mechanism and involved functional groups in metal-free and metal-loaded biomass of SRC for Pb(II) binding.
Similar content being viewed by others
Introduction
Industrialization and urbanization have resulted in a phenomenal increase in metallic contents in the environment and has emerged as a worldwide environmental problem. Heavy metals are innate constituents of the earth’s crust. Some are fundamental micronutrients for life, but at elevated concentrations they induce rigorous poisoning. Heavy metals are recalcitrant and in no way degrade in environment, but are only transformed and transferred (Satyawali et al. 2011; Hashim et al. 2011; Barka et al. 2013). Lead has no known biological functions. It is referred as a cumulative poison as it leads to biomagnifications at various trophic levels in food chains (Dauvin 2008; Flora et al. 2008; Lombardi et al. 2010). Lead is a mutagenic and teratogenic metal and induces stringent toxic effects such as cancer, hepatitis, neurodegenerative impairment, encephalopathy, renal failure, anaemia, and reproductive damage in living beings (Shahid et al. 2012; Ghazy and Gad 2014). Diverse industrial processes such as lead smelting, refining and manufacturing industries, battery manufacturing, printing and pigment, metal plating and finishing, ceramic and glass industries, and iron and steel manufacturing units are key sources of lead contamination in wastewater (Yurtsever and Sengil 2008; Sahu et al. 2013). Being a precarious neurotoxic metal, 10 µg/L of Pb(II) has been recommended by WHO as secure permissible level in drinking water (Watt et al. 2000; Naik and Dubey 2013). Inorganic form of lead is reviewed as a metabolic poison and enzyme inhibitor; still, organic forms of Pb(II) are extremely noxious (Anayurt et al. 2009; Javanbakht et al. 2011). Several methodologies such as chemical oxidation, electrocoagulation, electrodeposition, filtration, adsorption, chemical precipitation, solvent exchange, photo-degradation and membrane separation technologies have been explored long ago to mitigate recalcitrant heavy metals from adulterated wastewater (Verma et al. 2013; Rasool et al. 2013; Kumar et al. 2014; Zewaila and Yousef 2015). But these conventional methods proved unproductive due to their technical or economical constraints (Demir and Arisoy 2007; Cibati et al. 2013; Verma et al. 2015). Biological mechanisms impart a leading edge to current physico-chemical methods to wipe out noxious heavy metals from polluted waste water as they are eco-friendly, cost effective and do not produce colossal quantities of sludge (Cirik et al. 2013; Wang et al. 2014). Recently, biogenic sulphate reduction has been developing as an innovative bioprocess to remediate sulphate and heavy metals from effluents (Barrera et al. 2014; Lee et al. 2014; Sanchez-Andrea et al. 2014). Sulphate-reducing bacteria (SRB) metabolize organic matter in rigorous anaerobic environment using sulphate as an electron acceptor and subsequently results in generation of hydrogen sulphide and bicarbonate. This biogenic sulphide quickly reacts with heavy metal ions and finally transforms them into insoluble metal sulphides (Bratkova et al. 2013; Hao et al. 2014). SRB also possess other budding advantages such as they can reduce heavy metals directly by enzymatic approach and also endowed with high extracellular metal-binding capacity to accomplish bioremediation (Bridge et al. 1999; Pagnanelli et al. 2010; Sahinkaya et al. 2011; Wang et al. 2014). The present work aims at the study of influence of electron donors and optimization of bioprecipitation process to explore perfect conditions for efficient removal of Pb(II) and COD from simulated waste water. Therefore, the objective of current research is to present an effective method for treating Pb(II)-loaded wastewater with COD removal.
Materials and methods
Origin of sulphate-reducing consortium
The sulphate-reducing consortium (SRC) used in this research was derived from sludge of an electroplating industry SKH metals LTD., Manesar, Gurgaon district, Haryana, India. Sample was stored at anaerobic conditions in sealed plastic bag and was instantly brought to lab. From the sample, 5 g of sludge was added to 500 ml master culture flask filled with culture media containing modified Postgate media (g/L): Na2SO4 1.0; KH2PO4 0.5; NH4Cl 2.0; FeSO4 0.005; CaCl2 0.06; sodium citrate 0.3; yeast extract 0.1; sodium lactate 15 ml at pH 7 (Singh et al. 2011). Then master culture flask was purged with high-purity nitrogen to curtail the concentration of dissolved oxygen. The consortium was incubated for 2 months at 37 °C. The presence of black precipitates and foul odour of hydrogen sulphide was examined which indicates the presence of sulphate-reducing microbial consortia proficient in precipitating metal ions.
Experimental methods
Experiments were carried out in batch mode using 120-mL serum vials containing 100 mL of modified Postgate growth medium with pH 7 and were sealed with aluminum crimps and butyl rubber stoppers. After that 2 mL supernatant of SRC (5000 mg/L VSS) was injected in serum vials and were incubated at 37 °C in static position and were flushed with pure nitrogen gas (99 %) to set up anaerobic environment. For optimization experiments, incubation time was adjusted according to the experiments designed by Box–Behnken design model as shown in Table 2. Minimum inhibitory concentration (MIC) of Pb(II) ions for isolated sulphate-reducing bacterial consortium was analysed by implementing heavy metals tolerance assay. Lead nitrate (PbNO3)2; 1.598 g was dissolved in double distilled water (1 L) to obtain Pb(II) solution at 1000 mg/L. It was used as stock solution. Pb(II) varied in concentration ranging from 10 to 110 mg/L. Optical density (OD), metal removal (%) and protein content were studied to find out the minimum inhibitory concentration of Pb(II) ions. Different carbon sources like lactose, sucrose, glucose and sodium lactate were optimized using modified Postgate growth medium to ascertain best electron donor for anaerobic sulphate reduction. 3.0 % of total carbon content was supplemented from each carbon source. MIC and carbon source optimization experiments were performed at pH 7, temperature 37 °C and incubation time 7 days. All experiments were conducted in triplicate and average values were determined.
Diagnostic techniques
At defined time intervals, 5 mL of sample was collected from cultures using sterile and N2-purged syringe for analysis. OD, redox potential (Eh) and pH of withdrawn samples was measured instantly using EUTECH Instruments (pH and Eh Tutor). To prepare the cell-free supernatant, samples in vials were centrifuged at 5000 rpm for 10 min at 4 °C. Supernatant was further used for investigation of other parameters. The soluble chemical oxygen demand in the supernatant was measured by the Spectralab COD Digester (2015 M) and COD Titrator (CT-15) using the platinum combined electrode. Lowry’s method was performed to calculate protein content (Lowry et al. 1951; Bhatia et al. 2011) and OD was analysed at λ max 600 nm by UV–visible Spectrophotometer (T80 UV/VIS). Oxidation reduction potential (ORP) was monitored using pH 1500 Cyberscan (EUTECH Instruments). The ORP measurements as millivolt (mV) were carried out at room temperature. Total residual Pb(II) was quantified by atomic absorption spectrophotometer (Shimadzu AA-6300, Japan). Percent removal of Pb(II) was determined using the following equation:
where C i is the initial heavy metal concentration; C f is the final heavy metal concentration.
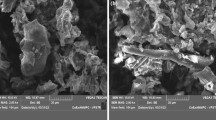
Morphology of Pb(II) sulphide precipitates was explored with scanning electron microscopy (SEM) using (JSM-E510LV, JEOL) in high-vacuum mode (accelerating voltage 10 kV) to study the morphology of metal loaded and unloaded biomass. The samples were prepared using phosphate buffer and 2 % glutaraldehyde and were kept overnight at 4 °C for fixation. Sodium cacodylate buffer (0.1 mol/L) was applied to wash the fixed granules and then samples were dewatered with a graded ethanol series, i.e. 10, 25, 50, 75, 90 and 100 %. Finally dewatered samples were dried to perform SEM.
Infrared analysis was accomplished to explain sorption mechanism to recognize the existing functional groups on SRC using PerkinElmer spectrum BX FTIR system (Beaconfield Buckinghamshire HP9 1QA) within range of 400–4000 cm−1 furnished with diffuse reflectance accessory. Fourier transform infrared (FTIR) measurements were carried out by the KBr technique. The samples were assorted with potassium bromide (KBr) and mounted beneath the spectrometer apparatus.
Box–Behnken design model
The optimization of biosorption process was performed using Box–Behnken design model (Bezerra et al. 2008) and was standardized on the basis of Design Expert software (Stat Ease, 9.0.4 trial version). In the present design, effect of individual variables, i.e. pH (5.0–9.0), temperature (32–42 °C) and time (5.0–9.0 days) were studied on responses, i.e. protein, Pb(II) precipitation and COD sequestration. Table 1 shows minimum and maximum level of each independent variable of the selected experimental design. Total 17 experiments were designed and conducted to study the simultaneous effect of these variables. The obtained responses and experiments run are shown in Table 2.
Statistical analysis
To predict the most favourable conditions for better elimination of Pb(II) and COD, quadratic equation [Eq. (2)] is expressed as follows: The quadratic equation model for predicting the optimal point was expressed according to Eq. (2).
where Yi (i = 3) is the predicted response, i.e. % removal of Pb(II) ions, COD and protein conc. (mg/mL), using SRC biomass, a o is constant coefficient, X i X j are the coded experimental variables correlated to response, e is the error of model and k is the number of variables studied (Nair and Ahammed 2015). The second-order polynomial Eq. (3), includes independent variables, coded as A, B and C and is expressed as follows:
In the present study, protein concentration (mg/mL), percentage removal of Pb(II) and COD were analysed using Eq. (3) including ANOVA to attain interaction between process variables and response. The quality-of-fit of polynomial model was expressed by the coefficient of determination r 2 and statistical significance was checked by F test in the program.
Results and discussion
Minimum inhibitory concentration (MIC) of Pb(II) ions
The minimum inhibitory concentration (MIC) of Pb(II) ions for SRC consortium was investigated using Postgate growth medium modified with varied concentration of heavy metal ranging from 10 to 110 mg/L. The cultures were incubated at 37 °C for a period of 7 days to monitor growth of SRB consortium. On 80 ppm concentration of Pb(II) ions, maximum OD, protein content and Pb(II) sequestration was found to be 0.98, 0.88 mg/mL and 99.8 %, respectively, as shown in Fig. 1. So MIC of Pb(II) ions for the said consortium was concluded to be 80 ppm.
Influence of electron donors on Pb(II) and COD removal
Sulphate-reducing bacterial consortium was cultured for Pb(II) removal. The modified Postgate growth medium was tested by varying different carbon sources such as lactose, sucrose, glucose and sodium lactate each with 3.0 % carbon content. Experiments were conducted to compare the pH profile, ORP (oxidation reduction potential), soluble COD removal and Pb(II) removal efficiency in presence of four carbon sources (Fig. 2). The lowest Pb(II) removal 59.2 and 70.3 % was obtained with lactose and sucrose, respectively. The substrate lactose, sucrose and glucose were fermented and assisted the fermentative bacteria as compared to sulphate-reducing bacteria. Thus fructose, sucrose and glucose resulted in reduction in pH of media and developed acidic conditions. This confirmed that fermentation had occurred when the consortium was supplemented with these carbon sources. The maximum Pb(II) removal achieved was 99.2 % with sodium lactate as carbon source. So it can be concluded that when sodium lactate was used as a carbon source at temperature 37 °C and time 7 days, maximum ORP, pH and soluble COD removal attained were −398, 8.06, and 55.5 %, respectively. So sodium lactate was considered to be the most efficient carbon source for the said sulphate-reducing bacterial consortium.
Optimization with response surface methodology
The interactive effect of different variables, i.e. pH (5.0–9.0), temperature (32–42 °C) and time (5–9 days) on responses, protein concentration (mg/mL), COD removal and bioprecipitation of Pb(II) with SRB consortium were studied by Box–Behnken design matrix and results are described in Table 3.
Validation of response surface models and statistical analysis
Data analysis by Box–Behnken design model reported optimum condition for COD removal and bioprecipitation of Pb(II) with SRB consortia and also examined the interactive effect of independent variables on the responses. On the basis of quadratic polynomial equations, correlation between the independent variables and responses were studied (4–6): the regression equation coefficients were calculated and data were fitted to a second-order polynomial equation to evaluate protein concentration (mg/mL), % COD removal and bioprecipitation of Pb(II) with SRB consortia.
The results of ANOVA for removal of protein concentration, COD and Pb(II) ions are mentioned in Table 3. Values of Prob > F less than 0.0001 designate that model terms are considerable for Pb(II) ions and COD sequestration. For this research the non-significant lack-of-fit (>0.05), is proficient for data fitness and demonstrated that quadratic model is quite satisfactory. In the experimental data, R-square (r 2) 0.9706 and adjusted r 2 0.9327 for protein, r 2 0.9928 and adjusted r 2 0.9835 for COD and r 2 0.9945 and adjusted r 2 0.9875 for Pb(II) is closer to 1.0 and rationalized the better fitness of model in the investigated data.
Optimization of variables for removal of Pb(II) and COD
The effect of independent variables pH (A), temperature (B) and contact time (C) on protein concentration, COD and Pb(II) removal with SRB consortia was studied using quadratic polynomial equations of response surface methodology (Eqs. (4)–(6)).
Independent variables, pH (A), temperature (B) and contact time (C), being crucial factors in bioprecipitation process, were studied intensively. As shown in Eqs. (4)–(6), it was analysed that all three independent variables have a linear positive effect on protein concentration, COD and Pb(II) elimination from aqueous solutions using SRB consortia.
First, pH (A) was an essential factor (P > 0.0001) and had a linear positive effect on protein concentration (mg/mL), biosorption of Pb(II) ions and COD removal from aqueous solution by SRB consortia (Eqs. (4)–(6)). It can be concluded that with increase in pH, protein concentration increases and also sequestration of COD and Pb(II) enhanced with rise in pH. As both chemical and biological factors affect Pb(II) sequestration, when sulphate-reducing bacteria (SRB) converts sulphate into sulphide, this biological process results in the generation of bicarbonate ions and increases the alkalinity of the medium which in turn provides the favourable conditions for SRB to develop and the sulphide then combines with heavy metal and results in formation of metal sulphide precipitates.
Second, temperature (B), (P > 0.0001) significantly influences COD and Pb(II) ions removal. Independent variable temperature had a positive effect on protein concentration (mg/mL), COD and Pb(II) elimination (Eqs. (4)–(6)). The increase in bioprecipitation of Pb(II) ions with increase in temperature is due increase in sulphate removal efficiency as concentration of protein is also increased with rise in temperature.
Third, incubation time (C), (P > 0.0001) also plays a fundamental role in removal of Pb(II) and had a positive effect on protein production, COD removal and Pb(II) precipitation by sulphate-reducing bacterial consortium (Eqs. (4)–(6)). As shorter HRT may not allow adequate time for SRB activity to neutralize acidity and precipitate metals. A longer HRT may imply depletion of either the available organic matter source or the sulphate source for SRB (Dvorak et al. 1992; Singh et al. 2011).
The interactive effect of two independent variables with another variable at a fixed level of protein concentration, biosorption of Pb(II) ions and COD removal with SRB consortium is shown in 3D surface plots (Figs. 3, 4, 5a–d).
Figure 3a–c shows the interactive effect of two variables pH (A) 5.0–9.0 and temperature (B) 32–42 °C on protein concentration, biosorption of Pb(II) ions and COD removal. In Fig. 3a, protein concentration initially increased with increase of pH i.e. up to 7 and then decreased with further increase of pH. Similar pattern was observed in case of temperature also. Maximum protein concentration was found to be 0.62 mg/mL at pH 7 and temperature 37 °C. SRBs mostly grow in neutral conditions of pH 6–8, but inhibition is detected at pH values below 6 or higher than 9 (Widdel 1988; Johnson et al. 2009; Zhao et al. 2011; Moon et al. 2013). In Fig. 3b, the removal of Pb(II) was first increased with increase in pH, i.e. up to 7 and then decreased with further increase of pH but slight variation was observed in precipitation of Pb(II) with change in temperature, i.e. from 32 to 42 °C. Hoa et al. (2007), revealed similar results as optimum pH was in range of 7.5–8.5 for lead sulphide precipitation through biological sulphate reduction process. Alvarez et al. (2007) reported similar findings as pH 7.5–8.0 was found to be the optimum pH for lead sulphide precipitation. In Fig. 3c, removal of COD was increased with increase of pH up to 7 and later it follows a decreasing trend with further increase of pH. Maximum removal of COD and Pb(II) was observed to be 91 and 98 %, respectively, at pH 7.0 and temperature 37 °C. It can be concluded from the results that maximum growth of SRB and maximum reduction in parameters were investigated at pH 7 and temperature 37 °C.
Figure 4a–c presents interactive effect of pH (A) and time (C) on concentration of protein, % removal of Pb(II) ions and COD with SRC. In Fig. 4a, protein concentration initially increased with increase in pH i.e. up to 7 and then decreased with further increase in pH. But in case of time, initially there was slight increase in protein concentration up to 7th day and further decreased with increase in time period. Maximum protein concentration was found to be 0.62 at pH 5 and time 7 days. Figure 4b shows that removal of Pb(II) ions was first increased and then decreased with increase of pH from 5.0 to 9.0 and no significant affect was perceived with increase of time, i.e. up to 5th to 9th day, In Fig. 4c, no significant effect was noticed in removal of COD with time 5–9 days while with pH, initially slight increase was observed in COD removal but later decreased with further increase in pH. Maximum removal of COD and Pb(II) was observed at pH 7.0 and incubation time 7 days. Wang et al. 2008 reported similar results as reducing rate is high when pH values are between 6 and 8 and lead removal rate was found to be >88.2 % when the pH value is 8.
Figure 5a–c shows interactive effect of two variables: time (C) 5.0–9.0 days and temperature (B) 32–42 °C on quantity of protein, sequestration of Pb(II) and removal of COD. In Fig. 5a concentration of protein initially increased with increase of temperature, i.e. up to 38 °C, and then decreased with further increase of temperature. Maximum protein concentration was found to be 0.62 mg/mL at temperature 38 °C and time 7 days. In Fig. 5b it is observed that no significant change was found in sequestration of Pb(II) with increase in temperature, i.e. 32–42 °C. While gradual increase was noticed in % removal of Pb(II) with increase in incubation time i.e. 5–9 days. Maximum Pb(II) ions removal was found to be 98 % at temperature 37 °C and time 7 days. In Fig. 5c, removal of COD was slightly increased with increase of temperature up to 37 °C and later it was almost stable with further increase of temperature. But with time COD removal followed an increasing trend with increase in incubation time, i.e. from 5 to 9 days. Maximum removal of COD was observed to be 91 % at pH 7.0 and time 7 days.
Characterization of Pb(II) sulphide precipitates
SEM was performed to further characterize the lead sulphide (PbS) precipitates from metal-free and metal-loaded biomass of SRC as shown in Fig. 6. SEM images revealed that the surface morphology of precipitates had a very muddled morphology of metal-loaded biomass with no definite pattern, as shown in Fig. 6b. Thus it can be inferred that there was profound effect of Pb(II) ions on surface of SRC biomass.
Fourier transform infrared spectroscopy (FTIR) study was carried out to identify the functional groups present in SRC in the range 4000–400 cm−1. The biosorbent capacity of SRC depends upon chemical reactivity of functional groups at the biomass surface. Figure 7 shows the shift in the wavelength of dominant peak associated with the plots by comparing between the control, i.e. lead-free and lead-loaded biomass. Metal binding process took place by precipitation of metal and also at the surface of SRC as shown by the shifts in the wavelength. Two peaks at 604.19 and 564.25 cm−1 disappeared and the peak at 872.28 cm−1 was flat showing that sulphate group was strongly involved in the adsorption of lead by adsorbent. The 1000–1400 cm−1 absorbtion band corresponds to –CH3, –CH2–, and C–F group, whereas the 750–1000 cm−1 band corresponds to S=O, –C–C–, and C–Cl functional groups.
Summary
In this study, MIC of Pb(II) ions was found to be 80 ppm. Using Box–Behnken design, it was concluded that combination of pH, temperature and incubation time, had significant effect on biosorption of Pb(II) precipitation and COD removal. The maximum responses were found to be 0.62 mg/mL protein concentration, 98 % Pb(II) and 91 % COD removal at optimum independent variables such as pH 7, temperature 37 °C and incubation time 7 days. From significant model and mathematical evaluation, this study deduced that RSM approach was found to be effective and efficient process for optimization of biosorption process.
References
Alvarez MT, Crespo C, Mattiasson B (2007) Precipitation of Zn(II), Cu(II) and Pb(II) at bench-scale using biogenic hydrogen sulfide from the utilization of volatile fatty acids. Chemosphere 66:1677–1683
Anayurt RA, Sari A, Tuzen M (2009) Equilibrium, thermodynamic and kinetic studies on biosorption of Pb(II) and Cd(II) from aqueous solution by macrofungus (Lactarius scrobiculatus) biomass. Chem Eng J 151:255–261
Barka N, Abdennouri M, Makhfouk ME, Qourzal S (2013) Biosorption characteristics of cadmium and lead onto eco-friendly dried cactus (Opuntia ficus indica) cladodes. J Environ Chem Eng 1:144–149
Barrera EL, Spanjers H, Romero O, Rosa E, Dewulf J (2014) Characterization of the sulfate reduction process in the anaerobic digestion of a very high strength and sulfate rich vinasse. Chem Eng J 248:383–393
Bezerra MA, Santelli RE, Oliveira EP, Villar LS, Escaleira LA (2008) Response surface methodology (RSM) as a tool for optimization in analytical chemistry. Talanta 76:965–977
Bhatia D, Kumar R, Singh R, Chadetrik R, Bishnoi NR (2011) Statistical modelling and optimization of substrate composition for bacterial growth and cadmium removal using response surface methodology. Ecol Eng 37:2076–2081
Bratkova S, Koumanova B, Beschkov V (2013) Biological treatment of mining wastewaters by fixed-bed bioreactors at high organic loading. Bioresour Technol 137:409–413
Bridge TAM, White C, Gadd GM (1999) Extracellular metal-binding activity of the sulphate-reducing bacterium Desulfococcus multivorans. Microbiology 145:2987–2995
Cibati A, Cheng KY, Morris C, Ginige MP, Sahinkaya E, Pagnanelli F, Kaksonen AH (2013) Selective precipitation of metals from synthetic spent refinery catalyst leach liquor with biogenic H2S produced in a lactate-fed anaerobic baffled reactor. Hydrometallurgy 139:154–161
Cirik K, Dursun N, Sahinkaya E, Cinar O (2013) Effect of electron donor source on the treatment of Cr(VI) containing textile wastewater using sulfate-reducing fluidized bed reactors (FBRs). Bioresour Technol 133:414–420
Dauvin JC (2008) Effects of heavy metal contamination on the macrobenthic fauna in estuaries: the case of the Seine estuary. Mar Pollut Bull 57:160–167
Demir A, Arisoy M (2007) Biological and chemical removal of Cr(VI) from waste water: cost and benefit analysis. J Hazard Mater 147:275–280
Dvorak DH, Hedin RS, Edenborn HM, McIntire PE (1992) Treatment of metal contaminated water using bacterial sulfate reduction: results from pilot-scale reactors. Biotechnol Bioeng 40:609–616
Flora SJS, Mittal M, Mehta A (2008) Heavy metal induced oxidative stress and its possible reversal by chelation therapy. Indian J Med Res 128:501–523
Ghazy SE, Gad AHM (2014) Lead separation by sorption onto powdered marble waste. Arabian J Chem 7:277–286
Hao TW, Xiang PY, Mackey HR, Chi K, Lu H, Chui HK, Loosdrecht MCMV, Chen GH (2014) A review of biological sulfate conversions in wastewater treatment. Water Resour 65:1–21
Hashim MA, Mukhopadhyay S, Sahu JN, Sengupta B (2011) Remediation technologies for heavy metal contaminated groundwater. J Environ Manage 92:2355–2388
Hoa TTH, Liamleam W, Annachhatre AP (2007) Lead removal through biological sulfate reduction process. Bioresour Technol 98:2538–2548
Javanbakht V, Zilouei H, Karimi K (2011) Lead biosorption by different morphologies of fungus Mucor indicus. Int Biodeterior Biodegrad 65:294–300
Johnson DB, Jameson E, Rowe OF, Wakeman K, Hallberg KB (2009) Sulfidogenesis at low pH by acidophilic bacteria and its potential for the selective recovery of transition metals from mine water. Adv Mater Res 71–73:693–696
Kumar N, Omoregie EO, Rose J, Masion A, Lloyd JR, Diels L, Bastiaens L (2014) Inhibition of sulfate reducing bacteria in aquifer sediment by iron nanoparticles. Water Res 51:64–72
Lee DJ, Liu X, Weng HL (2014) Sulfate and organic carbon removal by microbial fuel cell with sulfate-reducing bacteria and sulfide-oxidising bacteria anodic biofilm. Bioresour Technol 156:14–19
Lombardi PE, Peri SI, Verrengia NR (2010) ALA-D and ALA-D reactivated as biomarkers of lead contamination in the fish Prochilodus lineatus. Ecotoxicol Environ Saf 73:1704–1711
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the folin phenol reagent. J Biol Chem 193:265–275
Moon C, Singh R, Chaganti SR, Lalman JA (2013) Modeling sulfate removal by inhibited mesophilic mixed anaerobic communities using a statistical approach. Water Res 47:2341–2351
Naik MM, Dubey SK (2013) Lead resistant bacteria: lead resistance mechanisms, their applications in lead bioremediation and biomonitoring. Ecotoxicol Environ Saf 98:1–7
Nair AT, Ahammed MM (2015) The reuse of water treatment sludge as a coagulant for post-treatment of UASB reactor treating urban wastewater. J Clean Prod 96:272–281
Pagnanelli F, Viggi CC, Toro L (2010) Isolation and quantification of cadmium removal mechanisms in batch reactors inoculated by sulphate reducing bacteria: biosorption versus bioprecipitation. Bioresour Technol 101:2981–2987
Rasool K, Woo SH, Lee DS (2013) Simultaneous removal of COD and Direct Red 80 in a mixed anaerobic sulfate-reducing bacteria culture. Chem Eng J 223:611–616
Sahinkaya E, Gunes FM, Ucar D, Kaksonen AH (2011) Sulfidogenic fluidized bed treatment of real acid mine drainage water. Bioresour Technol 102:683–689
Sahu MK, Mandal S, Dash SS, Badhai P, Patel RK (2013) Removal of Pb(II) from aqueous solution by acid activated red mud. J Environ Chem Eng 1:1315–1324
Sanchez-Andrea I, Sanz JL, Bijmans MFM, Stams AJM (2014) Sulfate reduction at low pH to remediate acid mine drainage. J Hazard Mater 269:98–109
Satyawali Y, Seuntjens P, Van Roy S, Joris I, Vangeel S, Dejonghe W, Vanbroekhoven K (2011) The addition of organic carbon and nitrate affects reactive transport of heavy metals in sandy aquifers. J Contam Hydrol 123:83–93
Shahid M, Pinelli E, Dumat C (2012) Review of Pb availability and toxicity to plants in relation with metal speciation; role of synthetic and natural organic ligands. J Hazard Mater 219:1–12
Singh R, Kumar A, Kirrolia A, Kumar R, Yadav N, Bishnoi NR, Lohchab RK (2011) Removal of sulphate, COD and Cr(VI) in simulated and real wastewater by sulphate reducing bacteria enrichment in small bioreactor and FTIR study. Bioresour Technol 102:677–682
Verma A, Shalu Singh A, Bishnoi NR, Gupta A (2013) Biosorption of Cu (II) using free and immobilized biomass of Penicillium citrinum. Ecol Eng 61:486–490
Verma A, Dua R, Singh A, Bishnoi NR (2015) Biogenic sulfides for sequestration of Cr(VI), COD and sulfate from synthetic wastewater. Water Sci 29:19–25
Wang QL, Ding DX, Hue M, Yuran L, Qiu GZ (2008) Removal of SO4 2−, uranium and other heavy metal ions from simulated solution by sulfate reducing bacteria. Trans Nonferrous Met Soc China 18:1529–1532
Wang J, Li Q, Li MM, Chen TH, Zhou YF, Yue ZB (2014) Competitive adsorption of heavy metal by extracellular polymeric substances (EPS) extracted from sulfate reducing bacteria. Bioresour Technol 163:374–376
Watt GCM, Britton A, Gilmour HG, Moore MR, Murray GD, Robertson SJ (2000) Public health implications of new guidelines for lead in drinking water: a case study in an area with historically high water lead levels. Food Chem Toxicol 38:73–79
Widdel F (1988) Microbiology and ecology of sulfate-and sulfur-reducing bacteria. In: Zehnder AJB (ed) Biology of Anaerobic Microorganisms. Wiley Interscience, New York, pp 469–585
Yurtsever M, Sengil IA (2008) Biosorption of Pb(II) ions by modified quebracho tannin resin. J Hazard Mater 163:58–64
Zewaila TM, Yousef NS (2015) Kinetic study of heavy metal ions removal by ion exchange in batch conical air spouted bed. Alex Eng J 54:83–90
Zhao CQ, Yang QH, Chen WY, Li H, Zhang H (2011) Isolation of a sulfate reducing bacterium and its application in sulfate removal from tannery wastewater. Afr J Biotechnol 10:11966–11971
Acknowledgments
The author Anamika Verma is grateful to University Grant Commission, New Delhi, for awarding Basic Scientific Research (BSR) fellowship as Senior Research Fellowship (SRF) for this investigation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Verma, A., Bishnoi, N.R. & Gupta, A. Optimization study for Pb(II) and COD sequestration by consortium of sulphate-reducing bacteria. Appl Water Sci 7, 2309–2320 (2017). https://doi.org/10.1007/s13201-016-0402-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13201-016-0402-7