Abstract
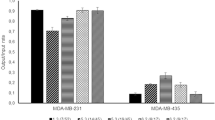
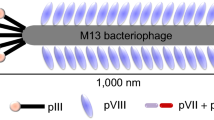
Cholangiocarcinoma (CCA) is a cancer of the bile duct with high mortality rate and poor prognosis, owing to the difficulty in the early diagnosis and prognosis. The specific biomarkers or affinity reagents toward CCA cells could be great tools to assist in detection of CCA. However, screening of biomarkers/affinity reagents are generally labor-intensive, time-consuming and requiring large volume of samples and reagents. Therefore, we developed an integrated microfluidic system which could automatically perform selections of biomarkers and affinity reagents using phage display techniques. The experimental results showed that the selection of phage-displayed peptides bound to CCA cells was successfully demonstrated on the integrated microfluidic system using fewer reagents, samples and less time (5.25 h per biopanning round, and continuously performed for only 4 panning rounds). Three oligopeptides were screened, and their specificity and affinity toward CCA cells were characterized. Furthermore, comparing to conventional EpiEnrich beads for cancer cell capture, the screened CCA-specific peptides showed relatively low capture rate over control normal cells. It is envisioned that this microfluidic system may be a powerful tool for screening of biomarkers/affinity reagents in clinical diagnosis and target therapy for CCA.







Similar content being viewed by others
Abbreviations
- bp:
-
Base pairs
- CA19-9:
-
Cancer antigen 19-9
- CCA:
-
Cholangiocarcinoma
- CCD:
-
Charge-coupled-device
- CEA:
-
Carcinoembryonic antigen
- CNC:
-
Computer-numerical-control
- DMEM:
-
Dulbecco’s modified eagle medium
- ddH2O:
-
Double-stilled water
- E. coli :
-
Escherichia coli
- EphA5:
-
Ephrin type-A receptor 5
- EphA2:
-
Ephrin type-A receptor 2
- EpiEnrich:
-
Epithelial Enrich
- FITC:
-
Fluorescein isothiocyanate
- FGFR2:
-
Fibroblast growth factor receptor 2
- FGFR3:
-
Fibroblast growth factor receptor 3
- FBS:
-
Fetal bovine serum
- IPTG/X-Gal:
-
Isopropyl β-D-1-thiogalactopyranoside/5-bromo-4-chloro-3-indolyl-d-galactoside
- LB:
-
Luria–Bertani
- NCKUH:
-
National Cheng Kung University Hospital
- PBS:
-
Phosphate buffer saline
- PCR:
-
Polymerase chain reaction
- PDMS:
-
Polydimethylsiloxane
- PMMA:
-
Polymethylmethacrylate
- RPMI:
-
Roswell park memorial institute
- SOC:
-
Super optimal broth with catabolite repression
References
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Anderson CD, Pinson CW, Berlin J, Chari RS (2004) Diagnosis and treatment of cholangiocarcinoma. Oncologist 9:43–57
Arai Y et al (2014) Fibroblast growth factor receptor 2 tyrosine kinase fusions define a unique molecular subtype of cholangiocarcinoma. Hepatology 59:1427–1434. doi:10.1002/hep.26890
Boot RG et al (2007) Identification of the non-lysosomal glucosylceramidase as β-glucosidase 2. J Biol Chem 282:1305–1312. doi:10.1074/jbc.M610544200
Bramucci E, Paiardini A, Bossa F, Pascarella S (2012) PyMod: sequence similarity searches, multiple sequence-structure alignments, and homology modeling within PyMOL. BMC Bioinf 13(Suppl 4):S2. doi:10.1186/1471-2105-13-S4-S2
Chang YH, Huang CJ, Lee GB (2012) A tunable microfluidic-based filter modulated by pneumatic pressure for separation of blood cells. Microfluid Nanofluid 12:85–94. doi:10.1007/s10404-011-0851-0
Che YJ, Wu HW, Hung LY, Liu CA, Chang HY, Wang K, Lee GB (2015) An integrated microfluidic system for screening of phage-displayed peptides specific to colon cancer cells and colon cancer stem cells. Biomicrofluidics 9:054121. doi:10.1063/1.4933067
Cui XD, Lee MJ, Kim JH, Hao PP, Liu L, Yu GR, Kim DG (2013) Activation of mammalian target of rapamycin complex 1 (mTORC1) and Raf/Pyk2 by growth factor-mediated Eph receptor 2 (EphA2) is required for cholangiocarcinoma growth and metastasis. Hepatology 57:2248–2260. doi:10.1002/hep.26253
Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A (2003) ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31:3784–3788
Haas SJ, Smith GP (1993) Rapid sequencing of viral DNA from filamentous bacteriophage. Biotechniques 15:422–424, 426–428, 431
Huang CW, Huang SB, Lee GB (2006) Pneumatic micropumps with serially connected actuation chambers. J Micromech Microeng 16:2265–2272. doi:10.1088/0960-1317/16/11/003
Ku JL et al (2002) Establishment and characterisation of six human biliary tract cancer cell lines. Br J Cancer 87:187–193. doi:10.1038/sj.bjc.6600440
Lavi A et al (2013) Detection of peptide-binding sites on protein surfaces: the first step toward the modeling and targeting of peptide-mediated interactions. Proteins 81:2096–2105. doi:10.1002/prot.24422
Liu Y, Adams JD, Turner K, Cochran FV, Gambhir SS, Soh HT (2009) Controlling the selection stringency of phage display using a microfluidic device. Lab Chip 9:1033–1036. doi:10.1039/b820985e
Maruyama M et al (2004) Establishment of a highly differentiated immortalized human cholangiocyte cell line with SV40T and hTERT. Transplantation 77:446–451. doi:10.1097/01.TP.0000110292.73873.25
Miyagiwa M, Ichida T, Tokiwa T, Sato J, Sasaki H (1989) A new human cholangiocellular carcinoma cell line (HuCC-T1) producing carbohydrate antigen 19/9 in serum-free medium. In Vitro Cell Dev Biol 25:503–510
Munz M et al (2010) Side-by-side analysis of five clinically tested anti-EpCAM monoclonal antibodies. Cancer Cell Int 10:44. doi:10.1186/1475-2867-10-44
Nesbit GM, Johnson CD, James EM, Maccarty RL, Nagorney DM, Bender CE (1988) Cholangiocarcinoma—diagnosis and evaluation of resectability by CT and sonography as procedures complementary to cholangiography. Am J Roentgenol 151:933–938
Pasquale EB (2010) Eph receptors and ephrins in cancer: bidirectional signalling and beyond. Nat Rev Cancer 10:165–180. doi:10.1038/nrc2806
Reinholz MM et al (2005) Evaluation of a panel of tumor markers for molecular detection of circulating cancer cells in women with suspected breast cancer. Clin Cancer Res 11:3722–3732. doi:10.1158/1078-0432.CCR-04-1483
Sefah K, Shangguan D, Xiong X, O’Donoghue MB, Tan W (2010) Development of DNA aptamers using Cell-SELEX. Nat Protoc 5:1169–1185. doi:10.1038/nprot.2010.66
Smith GP (1985) Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 228:1315–1317
Talasaz AH et al (2009) Isolating highly enriched populations of circulating epithelial cells and other rare cells from blood using a magnetic sweeper device. Proc Natl Acad Sci USA 106:3970–3975. doi:10.1073/pnas.0813188106
Tani K et al (1991) MR imaging of peripheral cholangiocarcinoma. J Comput Assist Tomogr 15:975–978
Van Beers BE (2008) Diagnosis of cholangiocarcinoma. HPB (Oxford) 10:87–93. doi:10.1080/13651820801992716
Wang J et al (2011) Selection of phage-displayed peptides on live adherent cells in microfluidic channels. Proc Natl Acad Sci USA 108:6909–6914. doi:10.1073/pnas.1014753108
Wang CH, Weng CH, Che YJ, Wang K, Lee GB (2015) Cancer cell-specific oligopeptides selected by an integrated microfluidic system from a phage display library for ovarian cancer diagnosis. Theranostics 5:431–442. doi:10.7150/thno.10891
Weng CH, Hsieh IS, Hung LY, Lin HI, Shiesh SC, Chen YL, Lee GB (2012) An automatic microfluidic system for rapid screening of cancer stem-like cell-specific aptamers. Microfluid Nanofluid 14:753–765. doi:10.1007/s10404-012-1095-3
Wong I, Ho CM (2009) Surface molecular property modifications for poly(dimethylsiloxane) (PDMS) based microfluidic devices. Microfluid Nanofluid 7:291–306. doi:10.1007/s10404-009-0443-4
Wu XR (2005) Urothelial tumorigenesis: a tale of divergent pathways. Nat Rev Cancer 5:713–725. doi:10.1038/nrc1697
Xu Y et al (2000) Multiple binding sites in collagen type I for the integrins α1β1 and α2β1. J Biol Chem 275:38981–38989. doi:10.1074/jbc.M007668200
Yang YN, Hsiung SK, Lee GB (2008) A pneumatic micropump incorporated with a normally closed valve capable of generating a high pumping rate and a high back pressure. Microfluid Nanofluid 6:823–833. doi:10.1007/s10404-008-0356-7
Yang SY, Cheng FY, Yeh CS, Lee GB (2010) Size-controlled synthesis of gold nanoparticles using a micro-mixing system. Microfluid Nanofluid 8:303–311. doi:10.1007/s10404-009-0461-2
Zhou MY, Gomez-Sanchez CE (2000) Universal TA cloning. Curr Issues Mol Biol 2:1–7
Acknowledgements
Authors would like to thank financial support from the National Health Research Institute in Taiwan (NHREI-EX104-10428EI) and Ministry of Science and Technology in Taiwan (MOST 105-2119-M-007-009).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yu, CW., Fu, CY., Hung, LY. et al. Screening of peptide specific to cholangiocarcinoma cancer cells using an integrated microfluidic system and phage display technology. Microfluid Nanofluid 21, 145 (2017). https://doi.org/10.1007/s10404-017-1983-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10404-017-1983-7