Abstract
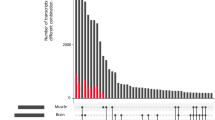
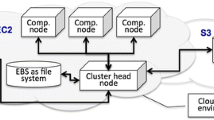
To determine annotations of the sequence elements on microarrays used for transcriptional profiling experiments in livestock species, currently researchers must either use the sparse direct annotations available for these species or create their own annotations. ANEXdb (http://www.anexdb.org) is an open-source web application that supports integrated access of two databases that house microarray expression (ExpressDB) and EST annotation (AnnotDB) data. The expression database currently supports storage and querying of Affymetrix-based expression data as well as retrieval of experiments in a form ready for NCBI-GEO submission; these services are available online. AnnotDB currently houses a novel assembly of approximately 1.6 million unique porcine-expressed sequence reads called the Iowa Porcine Assembly (IPA), which consists of 140,087 consensus sequences, the Iowa Tentative Consensus (ITC) sequences, and 103,888 singletons. The IPA has been annotated via transfer of information from homologs identified through sequence alignment to NCBI RefSeq. These annotated sequences have been mapped to the Affymetrix porcine array elements, providing annotation for 22,569 of the 23,937 (94%) porcine-specific probe sets, of which 19,253 (80%) are linked to an NCBI RefSeq entry. The ITC has also been mined for sequence variation, providing evidence for up to 202,383 SNPs, 62,048 deletions, and 958 insertions in porcine-expressed sequence. These results create a single location to obtain porcine annotation of and sequence variation in differently expressed genes in expression experiments, thus permitting possible identification of causal variants in such genes of interest. The ANEXdb application is open source and available from SourceForge.net.


Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (2000) Basic local alignment search tool. J Mol Biol 215:403–410
Bateman A, Coin L, Durbin R, Finn RD, Hollich V et al (2002) The Pfam protein families database. Nucleic Acids Res 30:276–280
Boguski MS, Lowe TM, Tolstoshev CM (1993) dbEST—database for “expressed sequence tags”. Nat Genet 4:332–333
Brett D, Lehmann G, Hanke J, Gross S, Reich J et al (2000) EST analysis online: WWW tools for detection of SNPs and alternative splice forms. Trends Genet 16:416–418
Cookson W, Lian L, Abecasis G, Moffatt M, Lathrop M (2009) Mapping complex disease traits with global gene expression. Nat Rev Genet 10:184–194
Ernst CW, Steibel JP, Ramos AM, Lunney JK, Wysocki M et al (2008) Assessment of the swine protein-annotated oligonucleotide microarray and utility of the arrays for eQTL and transcriptional profiling studies. Plant and Animal Genome XVI, San Diego, CA, 12–16 January 2008, pp W494
Gallant-Behm CL, Reno C, Tsao H, Hart DA (2007) Genetic involvement in skin wound healing and scarring in domestic pigs: assessment of molecular expression patterns in (Yorkshire × Red Duroc) × Yorkshire backcross animals. J Invest Dermatol 127:233–244
Gene Ontology Consortium (2000) Gene ontology: tool for the unification of biology. Nat Genet 25:25–29
Gentleman R, Carey VJ, Bates DM, Bolstad B, Dettling M et al (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5:R80
Gorbach DM, Hu ZL, Du ZQ, Rothschild MF (2009) SNP discovery in Litopenaeus vannamei with a new computational pipeline. Anim Genet 40:106–109
Gorodkin J, Cirera S, Hedegaard J, Gilchrist MJ, Panitz F et al (2007) Porcine transcriptome analysis based on 97 non-normalized cDNA libraries and assembly of 1,021,891 expressed sequence tags. Genome Biol 8:R45
Huang X, Madan A (1999) CAP3: a DNA sequence assembly program. Genome Res 9:868–877
Huntley D, Baldo A, Johri S, Sergot M (2006) SEAN: SNP prediction and display program utilizing EST sequence clusters. Bioinformatics 22:495–496
Kanehisa M, Goto S (2000) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28:27–30
Lai L, Prather RS (2002) Progress in producing knockout pigs for xenotransplantation by nuclear transfer. Ann Med 34:501–506
Lim D, Cho YM, Lee KT, Kang Y, Sung S et al (2009) The Pig Genome database (PiGenome): an integrated database for pig genome research. Mamm Genome 20:60–66
Mongan MA, Higgins M, Pine PS, Afshari C, Hamadeh H (2008) Assessment of repeated microarray experiments using mixed tissue RNA reference samples. Biotechniques 45:283–292
Nagaraj SH, Gasser RB, Ranganathan S (2006) A hitchhiker’s guide to expressed sequence tag (EST) analysis. Brief Bioinform 8:6–21
Panitz F, Stangaard H, Hornshøj H, Gorodkin J, Hedegaard J et al (2007) SNP mining porcine ESTs with MAVIANT, a novel tool for SNP evaluation and annotation. Bioinformatics 23:i387–i391
Picoult-Newberg L, Ideker TC, Pohl MG, Taylor SL, Donaldson MA et al (1999) Mining SNPs from EST databases. Genome Res 9:167–174
Pruitt KD, Tatusova T, Maglott DR (2007) NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts, and proteins. Nucleic Acids Res 35:D61–D65
Quackenbush J, Cho J, Lee D, Liang F, Holt I et al (2000) The TIGR Gene Indices: reconstruction and representation of expressed gene sequences. Nucleic Acids Res 28:141–145
Rogers CS, Stoltz DA, Meyerholz DK, Ostedgaard LS, Rokhlina T et al (2008) Disruption of the CFTR gene produces a model of cystic fibrosis in newborn pigs. Science 231:1837–1841
Schadt EE (2006) Novel integrative genomics strategies to identify genes for complex traits. Anim Genet 37:S18–S23
Sjölander K (2004) Phylogenomic inference of protein molecular function: advances and challenges. Bioinformatics 20:170–179
Slater G, Birney E (2005) Automated generation of heuristics for biological sequence comparison. BMC Bioinform 6:31–41
Tsai S, Cassady JP, Freking A, Nonneman DJ, Rohrer GA et al (2006) Annotation of the Affymetrix porcine genome microarray. Anim Genet 37:423
Uenishi H, Eguchi T, Suzuki K, Sawazaki T, Toki D et al (2007) PEDE (Pig EST Data Explorer) has been expanded into Pig Expression Data Explorer, including 10,147 porcine full-length cDNA sequences. Nucleic Acids Res 35:D650–D653
Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K et al (2008) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 36:D13–D21
Zhang Z, Schwartz S, Wagner L, Miller W (2000) A greedy algorithm for aligning DNA sequences. J Comput Biol 7:203–214
Zhu KQ, Carrougher GJ, Couture OP, Tuggle CK, Gibran NS et al (2008) Expression of collagen genes in the cones of skin in the Duroc/Yorkshire porcine model of fibroproliferative scarring. J Burn Care Res 29:815–827
Acknowledgments
We thank the USDA CSREES-NRI-2005-3560415618 and the ISU Center for Integrated Animal Genomics for funding this project. A USDA MGET 2001-52100-11506 Fellowship to O.C. is gratefully acknowledged. KC was funded under a NIH-NSF BBSI-0234102 award to Iowa State University.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Couture, O., Callenberg, K., Koul, N. et al. ANEXdb: an integrated animal ANnotation and microarray EXpression database. Mamm Genome 20, 768–777 (2009). https://doi.org/10.1007/s00335-009-9234-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00335-009-9234-1