Abstract
Ultraviolet (UV) radiation is a mutagen of major clinical importance in humans. UV-induced damage activates multiple signaling pathways, which initiate DNA repair, cell cycle arrest and apoptosis. To better understand these pathways, we studied the responses to UV-C light (254 nm) of germ cells in Caenorhabditis elegans. We found that UV activates the same cellular responses in worms as in mammalian cells. Both UV-induced apoptosis and cell cycle arrest were completely dependent on the p53 homolog CEP-1, the checkpoint proteins HUS-1 and CLK-2, and the checkpoint kinases CHK-2 and ATL-1 (the C. elegans homolog of ataxia telangiectasia and Rad3-related); ATM-1 (ataxia telangiectasia mutated-1) was also required, but only at low irradiation doses. Importantly, mutation of genes encoding nucleotide excision repair pathway components severely disrupted both apoptosis and cell cycle arrest, suggesting that these genes not only participate in repair, but also signal the presence of damage to downstream components of the UV response pathway that we delineate here. Our study suggests that whereas DNA damage response pathways are conserved in metazoans in their general outline, there is significant evolution in the relative importance of individual checkpoint genes in the response to specific types of DNA damage.
Similar content being viewed by others
Main
Maintenance of genome integrity is critically important for organisms to survive. In the presence of DNA damage, cells respond by activation of surveillance mechanisms that lead to cell cycle arrest and DNA repair. Highly damaged cells are often eliminated by apoptosis as a protective mechanism against the fixation of new mutations. The importance of these pathways is underscored by the observation that disruption of the DNA damage response pathway leads to an increased probability of cancer in humans.
In order to elucidate the complex molecular events that mediate the cellular responses to DNA damage, significant effort is being made in simple model organisms. The power of Caenorhabditis elegans as a system to genetically identify proteins that regulate apoptosis has been well established.1, 2 Recent analyses in C. elegans have identified a conserved checkpoint pathway that transduces the DNA damage signal caused by ionizing radiation (IR) to the cell cycle and the apoptotic machinery.3, 4 This pathway comprises the 9-1-1 complex (HUS-1/MRT-2/HPR-9) and a novel checkpoint protein, CLK-2, acting in parallel.5, 6 The CEP-1/p53 tumor suppressor protein is required for IR-induced apoptosis (but not cell cycle arrest). Activation of CEP-1/p53 leads to transcriptional upregulation of the BH3-only target genes, egl-1 and ced-13,5, 7, 8, 9 which in turn activate the C. elegans apoptotic machinery.1, 10
Ultraviolet (UV) light in the UV-B (<280 nm) and UV-C (280–315 nm) range damages DNA through the formation of cyclobutane pyrimidine dimers (CPDs) and 6-4 photoproducts (6-4 PPs).11, 12 UV radiation is a potent mutagen and both epidemiological and molecular evidence have established it as the major cause of human skin cancers. The removal of UV-induced DNA lesions is undertaken by the nucleotide excision repair (NER) machinery, which also removes adducts produced by chemotherapeutic agents, such as cisplatin.13 Two branches of NER, global genome repair (GGR) and transcription-coupled repair (TCR), have been identified. Both require multiple proteins that act in a multistep process: DNA damage recognition, DNA unwinding and incision, and finally repair synthesis and ligation.14, 15 The presence of UV-induced DNA lesions also activates members of the PI3 kinase family, ATR (ataxia telangiectasia and Rad3-related) and ATM (ataxia telangiectasia mutated).16, 17 ATR mediates the response to both UV- and IR-induced lesions as well as to stalled replication forks, whereas ATM is primarily responsible for the response to double-strand breaks (DSBs).
In this study, we describe our genetic analysis of the responses to UV-C light in the adult germ line of the nematode C. elegans. We delineate a signaling pathway that leads to cell cycle arrest and apoptosis. This pathway comprises several previously described components of the IR response pathway, including CEP-1/p53, HUS-1, CLK-2 and the C. elegans ATR homolog ATL-1. We also provide for the first time evidence that the checkpoint kinase CHK-2 and participates in DNA damage response in C. elegans. Importantly, we show that components of the NER machinery are required for UV-induced cell cycle arrest and apoptosis, and are necessary for the recruitment and activation of the 9-1-1 complex. Our results suggest that DNA damage response pathways evolved significantly during metazoan radiation, and confirm the usefulness of C. elegans as a model to dissect the responses to UV radiation in a multicellular organism.
Results
UV-C induces apoptosis in the C. elegans germ line
We previously showed that exposure of the adult C. elegans germ line to IR results in cell cycle arrest in the mitotic stem cell compartment and apoptotic death of pachytene cells.18 To determine the effects of UV light on germline physiology, which causes a pattern of DNA damage distinct from the one observed following IR,11 we exposed young adult hermaphrodites to increasing doses of UV-C radiation (254 nm) and measured the extent of germ cell apoptosis (Figure 1) and cell cycle arrest (see below) over time. We found that UV-C induces germ cell death in a dose-dependent manner, reaching a plateau at 100 J/m2 (Figure 1a). The kinetics of UV-C induced cell death were similar to those observed with IR, with increased apoptosis visible as early as 3 h post-treatment and persisting for at least 36 h (Figure 1b). These results indicate the presence in C. elegans of a signaling pathway that senses UV-C-induced damage, and conveys this information to the apoptotic machinery.
UV-C radiation induces apoptosis in the C. elegans germ line via transcriptional activation of egl-1 and ced-13. (a) Dose–response. Apoptotic germ cell corpses were scored in young adult animals 12 h after treatment with different doses of UV-C radiation. (b) Time course. Apoptotic germ cell corpses were scored at the indicated times following exposure to 100 J/m2 of UV-C radiation or 120 Gy of X-rays. (c) Transcriptional induction of egl-1 and ced-13 upon UV-C radiation. Relative mRNA levels of egl-1 and ced-13 were determined by real-time Q-RT-PCR in wild-type animals (wt), 12 h following treatment with either 120 Gy X-rays or 100 J/m2 UV-C. Data shown represent the average fold-change of three independent experiments±S.D. (d, e) Germ cell apoptosis following exposure to 100 J/m2 UV-C or 120 Gy X-rays was scored in egl-1(n1084n3082) (d) or ced-13(gk260) (e) mutants. Data shown represent the average of three independent experiments±S.D. (n>20 animals for each experiment)
UV-C-induced upregulation of egl-1 and ced-13 requires CEP-1/p53 and conserved checkpoint genes
To identify the genetic pathway that mediates UV-C-induced apoptosis, we concentrated our attention first on genes previously shown to mediate IR-induced apoptosis. An important step in IR-induced apoptosis in C. elegans is the CEP-1/p53-dependent transcriptional upregulation of egl-1 and ced-13, which encode proapoptotic BH3 domain proteins.5, 9 Using real-time Q-RT-PCR, we found that transcript levels of both egl-1 and ced-13 were also increased following UV treatment, albeit to a lesser extent than following IR (Figure 1c). Furthermore, loss of egl-1 and ced-13 function abrogated or reduced, respectively, UV-C-induced apoptosis (Figure 1d and e). Taken together, these observations suggest that UV-C induces apoptosis by stimulating increased expression of the BH3 domain proteins EGL-1 and CED-13.
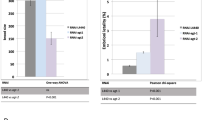
IR-induced cell death in the C. elegans germ line requires the p53 homolog, cep-1,7, 8 and the checkpoint genes hus-1, mrt-2 and rad-5/clk-2.4, 5, 6, 18 HUS-1 and MRT-2 are components of the 9-1-1 complex in C. elegans, whereas CLK-2 is the C. elegans homolog of Saccharomyces cerevisiae Tel2p. To investigate whether these well-conserved components also participate in the response to UV-C light, we quantified germline apoptosis following UV radiation in the respective mutant backgrounds. Loss of cep-1 and clk-2 completely abrogated UV-induced cell death (Figure 2a), whereas apoptosis was strongly reduced but not abolished in hus-1 or mrt-2 mutants (Figure 2b).
Genetic requirements for UV-C-induced apoptosis. Germ cell apoptosis following exposure to 100 J/m2 UV-C or 120 Gy X-rays was scored in (a) cep-1(gk138), clk-2(mn159), (b) hus-1(op244), mrt-2(e2663) and (c) chk-2(me64), chk-2(gk212) mutants. Data shown represent the average of three independent experiments±S.D. (n>20 animals for each experiment)
In mammals, the CHK2 kinase is known to activate p53 through phosphorylation on Ser2019 to induce apoptosis following exposure to IR and UV.20, 21 Surprisingly, C. elegans chk-2 mutants show a normal apoptotic response following IR.22 To determine whether CHK-2 might play a role in UV response, we analyzed chk-2 mutants for UV-induced germ cell apoptosis. Remarkably, both alleles of chk-2 that we tested, me64 (premature stop codon) and gk212 (large deletion; see Supplementary Figure 1c), failed to exhibit increased cell death upon UV-C treatment (Figure 2c). In contrast, IR caused an increase indistinguishable from wild type (Figure 2c), confirming the results of MacQueen and Villeneuve.22 Our findings are consistent with the hypothesis that UV-C induced phosphorylation of CEP-1 by CHK-2 leads to transcriptional activation of egl-1 and ced-13 and subsequent cell death.
The checkpoint kinase CHK1 has been implicated in UV-induced checkpoint activation in mammals.23 To determine the function of the C. elegans CHK1 ortholog CHK-1 in DNA damage response, we scored chk-1(RNAi) animals for apoptosis (chk-1 null mutants could not be tested, as they die during embryogenesis). Surprisingly, we found that chk-1(RNAi) animals were still proficient for both IR- and UV-C-induced germ cell apoptosis (Supplementary Figure 2). With the caveat that our analysis is based on RNAi, these results suggest that CHK-1 either plays a redundant role (perhaps together with CHK-2), or does not function at all in DNA damage-induced apoptosis.
ATL-1 is necessary to activate UV-induced germ cell apoptosis
In mammals, activation of CHK2 following DNA damage is mediated by the PI3K-like family members ATR and ATM.24, 25 Previous RNAi and genetic experiments have suggested that ATM-1 and ATL-1, the C. elegans orthologs of ATM and ATR, respectively, are required for apoptosis following IR.26, 27 To determine the role of these two kinases in the UV response pathway, we first analyzed germ cell apoptosis in animals carrying the null allele atl-1(tm853). We found that atl-1(tm853) mutants failed to induce germ cell apoptosis following treatment with either UV-C or IR (Figures 3a, b and 4a–d). Surprisingly, animals heterozygous for the tm853 deletion allele also showed a reduced response to both treatments, suggesting either that the tm853 deletion allele behaves as a dominant-negative mutation, or that the atl-1 locus is partially haploinsufficient for DNA damage-induced apoptosis. To distinguish between these two possibilities, we determined the response to UV-C of animals heterozygous for sDf29 or mDf1, two large genetic deficiencies that completely remove the atl-1 locus. Both sDf29/+ and mDf1/+ animals showed a reduced apoptotic response following UV (Figure 3c), supporting the latter hypothesis. In contrast to our observations, previous experiments with atl-1(tm853) showed that animals heterozygous for this allele have a wild-type response to IR-induced apoptosis.26 We do not know the cause for this discrepancy at the time being.
ATL-1 and ATM-1 are necessary for UV-C-induced egl-1 and ced-13 upregulation and germ cell apoptosis. (a, b) Time course of germ cell apoptosis in animals homozygous or heterozygous for the ATR deletion allele atl-1(tm853) following exposure to 100 J/m2 of UV-C radiation (a) or 120 Gy of X-rays (b). Data shown represent the average of three independent experiments±S.D. (c) Apoptotic germ cell corpses were scored 12 and 24 h post-exposure to 100 J/m2 of UV-C in young adult animals of the following genotypes: wild type (wt), atl-1(tm853), atl-1(tm853)/nT1, atl-1(tm853)/+, sDf29/+ and mDf1/+, the last two being large deficiencies that remove, among other, the atl-1 locus. Data shown represent the average of three independent experiments±S.D. (n>15 animals for each experiment). (d) DNA damage-induced germ cell apoptosis is compromised in atm-1(gk186) mutants after UV treatment, whereas it occurs normally upon X-ray irradiation. Young adult atm-1(gk186) animals were exposed to 100 J/m2 of UV-C radiation or 120 Gy of X-rays and germ cell corpses scored as described. (e) Transcriptional induction of egl-1 and ced-13 upon UV-C radiation is dependent on atl-1 and atm-1. Relative mRNA levels of egl-1 and ced-13 were determined by real-time Q-RT-PCR, 12 h following treatment with either 120 Gy of X-rays or 100 J/m2 of UV-C, in the indicated genetic backgrounds. Data shown represent the average fold-change of three independent experiments±S.D.
Apoptosis in response to low doses of DNA damage requires both atl-1 and atm-1. Apoptotic germ cell corpses were scored in the meiotic region of wild-type (wt) animals (a, b), atl-1(tm853) (c, d) and atm-1(gk186) (e, f) mutants after exposure to either 15 or 30 J/m2 of UV-C radiation (a, c, e) or 15 and 30 Gy of X-rays (b, d, f). Data shown represent the average of three independent experiments±S.D. (n>20 animals for each experiment). (g, h) Dose–response. Apoptotic germ cell corpses were scored in young adult wild-type (wt) or atm-1(gk186) animals 12 h after treatment with different doses of UV-C radiation (g) or X-rays (h). Data shown represent the average of three independent experiments±S.D. (n>20 animals for each experiment)
A dose-dependent requirement for ATM-1 in DNA damage-induced apoptosis
Whereas mammalian ATR responds primarily to UV or stalled replication forks, ATM reacts mainly to DSBs.17 To determine the function of C. elegans atm-1 in the DNA damage response, we characterized the behavior of atm-1(gk186) mutant animals. The gk186 mutation results in a 550 bp deletion in the 4.9 kb atm-1 ORF, which deletes parts of intron 1 and exon 2 (Supplementary Figure 1a). RT-PCR analysis revealed the presence of several truncated mRNA species in gk186 mutants (Supplementary Figure 1a); sequencing of the major mRNA product revealed that the splicing machinery used a cryptic splice acceptor site within exon 2, resulting in a frame shift and the premature termination of translation. As all conserved domains are either within or downstream of the gk186 deletion, we suspect that the mutant is a functional null.
The atm-1(gk186) mutants showed a strong induction of germ cell apoptosis following 120 Gy of X-rays (Figure 3d). However, the response to lower doses of IR (15 or 30 Gy) was seriously compromised (Figure 4f). In contrast, the apoptotic response of atm-1(gk186) mutants to UV-C was significantly reduced at both high and low doses: death induction was reduced by more than 50% at 100 J/m2 (Figure 3d), and appeared completely absent at 15 J/m2 (Figure 4e). Dose–response studies confirmed these distinct response patterns (Figure 4g and h).
Our results suggest that, under low damage conditions, ATM-1 function is crucial for the induction of apoptosis following UV-C or IR. The reduced requirement for ATM-1 at higher doses of IR could be due to activation or recruitment of additional proteins or pathways that act redundantly with, or bypass ATM-1.
Where in the signaling pathway do ATL-1 and ATM-1 act? To determine whether atl-1 or atm-1 function is required for UV-induced transcriptional upregulation of egl-1 and ced-13, we measured mRNA levels of these proapoptotic genes following DNA damage in atl-1(tm853) and atm-1(gk186) mutants (Figure 3e). Consistent with the apoptosis defect, we described above, upregulation of both genes was strongly impaired in atl-1 mutants. The response pattern of atm-1(gk186) mutants was more complex: ced-13 induction was significantly reduced following UV-C, whereas egl-1 levels remain largely unaffected. These results indicate that ATL-1 and ATM-1 function upstream of egl-1 and ced-13 and suggest that ATM-1 might be preferentially required for upregulation of ced-13 in response to UV-C.
The NER machinery is required for triggering UV-C-induced apoptosis
UV-induced DNA damage is normally recognized and repaired via the NER pathway in eukaryotes. We posited that recognition of UV lesions by the NER machinery might be required not only for DNA repair, but also to activate the downstream signaling pathways that lead to apoptosis. To test this hypothesis, we analyzed the effect on UV-C-induced apoptosis of two previously uncharacterized mutations in the NER genes xpc-1(Y76B12C.2) and xpa-1,28 which encode the C. elegans homologs of xeroderma pigmentosum complementation group C (XPC) and xeroderma pigmentosum complementation group A (XPA), respectively.
Animals homozygous for the xpc-1(ok734) mutation showed reduced apoptosis following UV-C treatment, whereas their response to IR was normal (Figure 5a). Surprisingly, we found that the ok734 mutation removes 1.7 kb of intron 3, but does not affect any exonic sequences (Supplementary Figure 1d). Indeed, a normal-size transcript could still be detected in ok734 mutants, likely explaining the mild defect that we observed. Consistent with this hypothesis, animals treated with xpc-1(RNAi) resulted in a much more severe defect (Figure 5b), suggesting that XPC function is necessary for UV-induced apoptosis.
The NER machinery is required for triggering UV-C-induced germ cell apoptosis. Staged xpc-1(ok734) (a), xpc-1(RNAi) (b) and xpa-1(ok698) (c) young adults were treated with either 100 J/m2 UV-C or 120 Gy X-rays and apoptotic germ cell corpses were scored at the indicated time points. (d, e) Germ cell apoptosis was scored every 12 h until 48 h post L4/adult molt in staged: xpa-1(ok698) and xpa-1(ok698) cep-1(RNAi) (d) or atm-1(gk186) xpa-1(ok698) (e) animals. Data shown represent the average of three independent experiments±S.D. (n>20 animals for each experiment)
The xpa-1(ok698) deletes half of the ORF, and leads to a degradation of the deletion transcript (Supplementary Figure 1e). Thus, this mutation is likely to be a null. Koo and co-workers previously reported that xpa-1(RNAi) animals exhibit reduced embryonic survival and survival to adulthood rate upon UV-C.29 We found that UV-induced apoptosis is completely abolished in the absence of xpa-1 (Figure 5c), consistent with the hypothesis that lack of proper damage recognition can disturb signaling to the apoptotic machinery. In contrast to UV, IR-induced apoptosis still occurs in the absence of xpa-1, although with somewhat reduced levels compared with the wild type (Figure 5c). The slight defect that we observed implies that a fraction of the DNA damage generated by X-rays likely is recognized and repaired via the NER pathway.
Notably, non-UV-treated xpc-1(RNAi) and xpa-1(ok698) animals had more germ cell apoptosis than wild-type worms (Figure 5b and c). We postulate that the accumulation of unrepaired endogenous damage in these animals eventually leads to apoptosis through activation of (an)other DNA damage signaling pathway(s). Indeed, the increased basal germ cell apoptosis was abrogated in xpa-1(ok698); cep-1 (RNAi) and atm-1(gk186) xpa-1(ok698) animals (Figure 5d and e), suggesting that this increase was due to activation of an atm-1- and cep-1-dependent pathway.
The HUS-1 complex acts downstream of NER to promote UV-C-induced apoptosis
The results we presented above suggest that recognition and/or processing of DNA damage by the NER machinery is an essential, and likely early step in UV-C-induced apoptosis. To more precisely position the NER components within this signaling cascade, we determined the subcellular distribution of the 9-1-1 complex subunit HUS-1 in some of the mutants we described above.
We previously showed that HUS-1∷GFP, which is normally diffusely distributed in the nuclei of germ cells, accumulates into distinct foci upon IR.5 We found that HUS-1∷GFP also formed foci following exposure to UV-C light (Figure 6a and b), consistent with our observation that HUS-1 participates in UV-C-induced apoptosis (Figure 2b). Elimination of xpa-1 (which is required for apoptosis in response to UV-C but not IR) dramatically reduced the foci accumulation following UV treatment (Figure 6a and b). By contrast, foci formation following IR was still robust (Figure 6b). These results suggest that the NER proteins act upstream of 9-1-1 complex recruitment.
UV-C-induced germ cell apoptosis requires recruitment of HUS-1 downstream of NER. (a) Fluorescent microscopy of mid-late pachytene germ cells expressing HUS-1∷GFP (opIs34). Germ cell nuclei from staged young adult wild-type (wt) or xpa-1(ok698) hermaphrodites expressing HUS-1∷GFP were scored for the presence of GFP, 4 h after exposure to 120 Gy of X-rays or 100 J/m2 of UV-C. Relocalized HUS-1 is seen as bright foci marked by arrowheads, whereas it is diffusely distributed before treatment. (b) UV-C-induced HUS-1 foci require XPA-1. Quantification of the GFP-positive signals shown in (a) following treatment with IR or UV-C in wild-type, xpa-1(ok698), atm-1(gk186) and cep-1(RNAi)-treated animals
We also analyzed HUS-1∷GFP recruitment in atm-1 and cep-1 mutants. We found that the basal levels of HUS-1∷GFP foci was elevated in the absence of atm-1, suggesting elevated levels of endogenous DNA damage and/or decreased repair activity in the mutants (Figure 6b). Both IR and UV treatment further increased the number of HUS-1∷GFP foci, indicating that ATM-1 is not required for the recruitment of the 9-1-1 complex. Similarly, knockdown of cep-1 did not significantly alter HUS-1∷GFP foci formation following DNA damage (Figure 6b). Thus, ATM-1 and CEP-1 likely act either downstream of, or in parallel to 9-1-1 complex recruitment.
Together, our findings suggest that recruitment of the HUS-1-containing 9-1-1 complex is likely a critical step in the UV-C response pathway, which occurs downstream of the action of the NER machinery, and which in turn is required for ATM-1- and CEP-1-dependent apoptosis and cell cycle arrest.
Cell cycle arrest upon UV-C requires a distinct set of genes that partially overlaps with that activated upon IR
We previously showed that IR treatment of C. elegans germ cells induced not only apoptosis but also cell cycle arrest of the mitotic stem cells present at the distal end of the germ line.18 To determine whether UV treatment also interferes with germ cell proliferation, we measured the number of nuclei present at the distal end of the gonad. We found that UV-C induces a transient proliferation arrest in mitotic germ cells. Wild-type animals showed a detectable decrease in cell numbers already 4 h post-treatment that lasted for at least 12 h, but which recovered by 24 h post-treatment (Figure 7a). To confirm that the reduced number of mitotic germ cells was caused by a proliferation arrest, we quantified the number of germ cells in mitosis following DNA damage. We found that arrested mitotic germ cell compartments showed a reduced mitotic index, based both on DAPI staining and probing of fixed germ lines with an antibody that specifically recognizes phospho-Ser10 H3, which is a marker of mitosis (Supplementary Figure 3).
UV-C irradiation causes cell cycle arrest in the mitotic zone of the germ line. (a) Staged L4 wild-type animals or cep-1(gk138) mutants were irradiated with 100 J/m2 UV-C. The number of germ cell nuclei present within the distal most 75 μm of the germ line was determined in a time course. (b) Staged L4 animals of the following genotypes were irradiated with 100 J/m2 UV-C or 120 Gy X-rays: wild-type (wt), hus-1(op244), clk-2(mn159), cep-1(gk138), chk-2(gk212), atl-1(tm853), atm-1(gk186) and xpa-1(ok698). The number of germ cell nuclei was determined as above 8 h after treatment. White bars represent the non-treated and light/dark gray bars the UV-C/IR-treated condition. Data shown represent the average from 40 gonads±S.E.M.
To determine the genetic requirements for UV-C-induced cell cycle arrest, we analyzed the cell cycle phenotype of the mutants we described above. We found that all genes required for UV-C-induced apoptosis also participate in UV-C-induced cell cycle arrest (Figure 7b). The relative strengths of the apoptosis and cell cycle arrest defects observed in the various mutants correlated well, with the exception of atl-1 (tm853), which showed a strong loss of apoptosis but only a partial defect in cell cycle arrest. Interestingly, the pathway that induces cell cycle arrest in response to UV-C overlaps only partially with the IR response pathway that we and others described previously.4, 27 Whereas hus-1, clk-2 and atl-1 are clearly required in both situations, cep-1, chk-2, atm-1 and the NER gene xpa-1 are required only for UV response (Figure 7b).
NER and checkpoint proteins are required for efficient repair of UV-C-induced DNA damage
To assess the capacity of our various mutants to repair UV-C-induced lesions, we measured the survival of embryos generated from UV-treated germline nuclei. xpa-1(ok698) mutant animals showed the most severe reduction in survival following UV-C treatment, consistent with the predicted essential role of xpa-1 in NER (Supplementary Figure 4). Additionally, their somatic tissues showed a strong sensitivity by arresting their development, when younger animals were subjected to UV radiation (see Materials and methods). atm-1(gk186) mutants displayed slightly decreased embryonic survival already under normal growth conditions, as did the checkpoint mutants hus-1(op244) and clk-2(mn159) (Supplementary Figure 4). This, together with the increased incidence of HUS-1∷GFP foci in atm-1 mutants (Figure 6), supports the notion that ATM-1 plays an important role in promoting the repair of endogenous DNA damage. Following treatment with UV-C, we observed a further reduction in survival in all three mutants, suggesting that repair of UV-C lesions is slightly defective or slowed down in all three cases. Interestingly, both atm-1 hus-1 and atm-1; clk-2 double mutants showed a strong synthetic lethal phenotype, reaching levels of survival as low as 20 and 50%, respectively (Supplementary Figure 4). A similar synthetic phenotype was described previously in hus-1; clk-2 double mutants.6 These observations imply the existence of significant redundancy in the pathway(s) that mediate repair of endogenous DNA damage in the C. elegans germ line.
Interestingly, although cep-1 animals are strongly defective in UV-induced apoptosis and cell cycle arrest, these mutants showed survival levels similar to the wild type (Supplementary Figure 4). This observation leads to two conclusions. First, CEP-1 is not required for DNA repair in response to UV-C. Second, and rather surprisingly, lack of apoptosis or cell cycle arrest does not appear to increase the proportion of cells that generate inviable embryos, at least under our experimental conditions. More precise assays will be required to determine the exact contribution of DNA damage-induced cell cycle arrest and apoptosis to the maintenance of genome stability.
Discussion
In this paper, we describe our genetic analysis of the cellular responses to UV-C light in the adult C. elegans germ line. We showed, for the first time, that UV-C induces both apoptotic cell death of meiotic cells and cell cycle arrest of the proliferating mitotic stem cells. We identified over half a dozen genes required for both responses. These genes act in a signaling pathway that overlaps with, but is distinct from the pathway that is activated in response to IR4 (Figure 8).
A genetic pathway for UV-C-induced apoptosis and cell cycle arrest. (a) Based on our genetic studies, our experiments with the hus-1∷gfp reporter, and data from other species we propose that UV-C-induced damages are recognized by NER pathway components, which subsequently activate, directly or indirectly, the 9-1-1 complex. The 9-1-1 complex in turn promotes activation of the checkpoint kinase cascade, leading to p53-dependent apoptosis and cell cycle arrest. The requirement for some of the UV-C pathway components is different in response to IR-induced damages (b and see text)
Importantly, we found that the NER pathway, which is implicated in the recognition and repair of UV-C-induced lesions (pyrimidine dimers and 6-4 PPs), also plays an essential role in activating cell cycle arrest and apoptosis. It is possible that assembly of the NER recognition complex at a site of damage is by itself sufficient to activate the signaling cascade. Alternatively, signaling could be activated upon recruitment of the repair machinery, or by the repair process itself (e.g., through the generation of regions of ssDNA). Further analysis of mutants defective in later steps in NER might allow us to distinguish between these various models. Interestingly, Cimprich and co-workers recently showed that XPA, but not other components of the NER pathway, is required for ATR activation in mammalian cell culture following UV damage.30 Unfortunately, the authors did not comment on whether the effect on ATR activation correlated with any change in apoptosis or cell cycle arrest in their experimental system.
Indeed, whereas mammalian cells defective in various NER components (e.g., the various XP complementation groups) often do show altered responses to UV light, the patterns of changes tend to be more complicated than the ones we observed in C. elegans: depending on the specific mutation and the gene affected, defective cells can either show increased or decreased levels of UV-induced apoptosis and/or cell cycle arrest.31, 32, 33, 34 It is likely that the complex response pattern observed in mammals in response to UV simply reflects the presence of multiple apoptosis-inducing pathways in these species. For example, unrepaired UV-induced lesions in ERCC1 or ERCC3-deficient (murine XP genes) cells do not activate the apoptotic machinery directly, but rather are converted into other types of damage (e.g., dsDNA strand breaks) during subsequent S phases, which in turn lead to apoptosis.33 The presence of these additional pathways might have precluded identification of the role of the NER pathway in promoting apoptosis in mammals.
The greater simplicity of the C. elegans DNA damage response network is also underscored through our analysis of the 9-1-1 complex subunit HUS-1. Whereas C. elegans hus-1 mutants are completely defective in both UV and IR-induced apoptosis and cell cycle arrest5, 18 (Figure 2b), implicating the 9-1-1 complex as an essential component in both pathways, mouse hus-1−/− cells in fact show increased apoptosis in response to IR.35, 36
What is the exact function of the 9-1-1 complex in UV-C-induced apoptosis? We found that UV-C irradiation led to a significant increase in HUS-1∷GFP foci (Figure 6); these foci are thought to represent either sites of persistent damage or of active repair.5 Interestingly, the 9-1-1 complex is thought to be recruited in response to dsDNA breaks. HUS-1∷GFP foci were not increased in xpa-1 mutants, indicating that activation of NER is required, directly or indirectly, for 9-1-1 complex recruitment. One possibility is that NER generates as a by-product a significant number of dsDNA breaks, perhaps when damaged sites on opposite strands are repaired simultaneously.
In contrast to the situation in xpa-1 mutants, HUS-1∷GFP foci still formed upon UV treatment in atm-1 and cep-1 mutants. Thus, these genes act either in parallel to or downstream of the 9-1-1 complex (Figure 8). Further biochemical studies will be required to dissect in more detail the exact order of molecular events in this signaling cascade. The strong synthetic lethality that we observed in atm-1 hus-1 double mutants suggests that these two proteins do have some non-overlapping activities, at least with respect to promotion of DNA repair.
In mammals, the checkpoint kinase CHK2 plays an important role in promoting apoptosis and cell cycle arrest in response to a variety of DNA damaging agents, including IR, UV and various mutagens and chemotherapeutic agents.37 Surprisingly, chk-2 mutants in C. elegans show a normal response to IR,22 which raised the issue of whether chk-2 might have a different function in invertebrates. We now showed that chk-2 mutants are strongly defective in both apoptosis and cell cycle arrest following UV treatment (Figures 2c and 7b). Thus, the molecular function of CHK-2 is likely conserved also in C. elegans, although the signals to which it responds appear likely to be distinct in worms and mammals.
We also made a similar observation with ATM-1 and ATL-1, the orthologs of ATM and ATR, respectively. In mammals, ATM mainly mediates response to dsDNA breaks, whereas ATR is activated by UV and stalled replication forks.16, 17 In C. elegans, we found that ATL-1 was the more important player in both UV- and IR-response pathways, with loss of ATM-1 only showing a strong defect at low doses27 (Figures 3a, d and 4c–f). Finally, we found a differential requirement for CEP-1/p53 in UV- and IR-induced cell cycle arrest: whereas mitotic germ cells in cep-1 mutants showed a normal response following IR treatment, they failed to arrest in response to UV light (Figure 7a). We surmise that p53 either is not activated in response to IR or that it acts redundantly with another signaling protein under these conditions.
Taken together, our observations suggest that there is a certain flexibility for organisms to change, over evolutionary time, the types of damage a given checkpoint protein responds to. This flexibility might be important in allowing organisms to adapt to new challenges as they change their life history (e.g., longer lifespans) or ecological niches.
Materials and Methods
Genetics
All strains were grown at 20°C on NGM agar seeded with Escherichia coli OP50.38 The Bristol N2 strain was used as the wild-type strain. The following mutations were used: LGI: atm-1(gk186), hus-1(op244), xpa-1(ok698), cep-1(gk138); LGIII: clk-2(mn159), mrt-2(e2663); LGIV: xpc-1(ok734); LGV: atl-1(tm853), sDf29, mDf1, egl-1(n1084n3082), chk-2(me64), chk-2(gk212); LGX: ced-13(gk260). Essential mutations were maintained as balanced strains: atl-1(tm853)/nT1[qIs51](IV;V), dpy-18(e364)/eT1 III; unc-46(e177) sDf29/eT1 V, dpy-18(e364)/eT1 III; mDf1/eT1 V and chk-2(me64) rol-9(sc148)/unc-51(e369) rol-9(sc148).
Germline apoptosis
Staged young adult worms (12 h post the L4/adult molt) were exposed to different doses of UV-C light (254 nm) (J/m2) or X-rays (Gy). A Stratalinker UV crosslinker, model 1800 (Stratagene) and an Isovolt 160/225/320/450 HS X-ray machine (Rich. Seifert & Co.) were used to deliver the appropriate doses. Corpses were scored in the meiotic region of one gonad arm at indicated time points using Nomarski optics, as described by Gumienny et al.39 For the RNAi experiments, staged L1 larvae were transferred onto plates seeded with bacteria expressing the respective RNAi clone40 and scored as young adults for germline apoptosis.
Relative quantification of transcripts
Staged young adults were treated with UV-C light or X-rays as described above. Animals were selected for processing 12 h post-treatment. Total RNA extraction, cDNA synthesis and quantitative real-time RT-PCR were performed as described previously.5 Transcript levels of egl-1 and ced-13 were normalized to 18S rRNA, tbp-1 and pgk-1 mRNAs, which served as internal controls. The forward and reverse primer sequences for tbp-1 and pgk-1 were: 5′-TTGGATTTGAAGAAGATTGCATTG-3′, 5′-AATGACTGCTGCGAAACGTTT-3′ and 5′-GCGATATTTATGTCAATGATGCTTTC-3′, 5′-TGAGTGCTCGACTCCAACCA-3′, respectively. Primer sequences for the other genes were described in Hofmann et al.5
Cell cycle arrest studies
Staged L4 larvae were treated with either 100 J/m2 of UV-C light or 120 Gy of X-rays. The cell cycle arrest phenotype was assessed at indicated time points by counting the number of mitotic nuclei present in one focal plane within 75 μm of the distal tip cell. Alternatively, dissected gonads were stained with an antibody against a phosphorylated form of histone H3 (anti-phospho-H3 (Ser10)) and DAPI, 7 h after treatment. DIC images were captured in both cases using an ORCA-ER digital CCD camera and analyzed using Openlab software.
Immunocytochemistry
For antibody staining of gonads, L4 hermaphrodites were dissected and fixed in 3% para-formaldehyde/0.1 M K2HPO4 (pH 7.2) for 50 min at room temperature, followed by a 10 min-incubation in 100% methanol on ice. Gonads were blocked in 5% BSA/PBS-Tween-20 0.1% for 1 h, followed by incubation with 1 : 100 anti-phospho-histone H3 polyclonal antibody (Ser10) (Upstate) overnight at 4°C. Alexa fluor 594 goat anti-rabbit IgG (Molecular Probes) was used as secondary antibody (1 : 500). The tissues were co-stained with DAPI before mounting. Fluorescent images were captured with a Leica DMRA2 microscope equipped with an ORCA-ER digital CCD camera and were processed with Openlab software.
Embryonic survival assay
Staged L4 larvae were subjected to 100 J/m2 UV-C. After 24 h recovery period, animals were singly transferred to new seeded plates (n≈50) and were left to lay eggs for 4–6 h (n⩾30 embryos/plate). As an exception, due to the strong sensitivity of the somatic tissues to the effects of UV-C, xpa-1(ok698) and xpc-1(ok734) mutants were treated 12 h post the L4 stage. Adults were then removed and the number of eggs laid was determined. Unhatched eggs were scored 24 h later to calculate the % embryonic survival. Moreover, owing to the fully penetrant maternal-effect lethal phenotype of atl-1(tm853) mutants, we did not include them in our assay.
Abbreviations
- UV:
-
ultraviolet light
- IR:
-
ionizing radiation
- ATM:
-
ataxia telangiectasia mutated
- ATR:
-
ataxia telangiectasia and Rad3-related
- CHK1/2:
-
checkpoint kinase 1/2
- CPDs:
-
cyclobutane pyrimidine dimers
- 6-4 PPs:
-
6-4 photoproducts
- NER:
-
nucleotide excision repair
- GGR:
-
global genome repair
- TCR:
-
transcription-coupled repair
- DSBs:
-
double-strand breaks
- XPA:
-
xeroderma pigmentosum complementation group A
- XPC:
-
xeroderma pigmentosum complementation group C
References
Metzstein MM, Stanfield GM, Horvitz HR . Genetics of programmed cell death in C. elegans: past, present and future. Trends Genet 1998; 14: 410–416.
Kinchen JM, Hengartner MO . Tales of cannibalism, suicide, and murder: programmed cell death in C. elegans. Curr Top Dev Biol 2005; 65: 1–45.
Hofmann ER, Milstein S, Hengartner MO . DNA-damage-induced checkpoint pathways in the nematode Caenorhabditis elegans. Cold Spring Harb Symp Quant Biol 2000; 65: 467–473.
Stergiou L, Hengartner MO . Death and more: DNA damage response pathways in the nematode C. elegans. Cell Death Differ 2004; 11: 21–28.
Hofmann ER, Milstein S, Boulton SJ, Ye M, Hofmann JJ, Stergiou L et al. Caenorhabditis elegans HUS-1 is a DNA damage checkpoint protein required for genome stability and EGL-1-mediated apoptosis. Curr Biol 2002; 12: 1908–1918.
Ahmed S, Alpi A, Hengartner MO, Gartner A . C. elegans RAD-5/CLK-2 defines a new DNA damage checkpoint protein. Curr Biol 2001; 11: 1934–1944.
Derry WB, Putzke AP, Rothman JH . Caenorhabditis elegans p53: role in apoptosis, meiosis, and stress resistance. Science 2001; 294: 591–595.
Schumacher B, Hofmann K, Boulton S, Gartner A . The C. elegans homolog of the p53 tumor suppressor is required for DNA damage-induced apoptosis. Curr Biol 2001; 11: 1722–1727.
Yan N, Gu L, Kokel D, Chai J, Li W, Han A et al. Structural, biochemical, and functional analyses of CED-9 recognition by the proapoptotic proteins EGL-1 and CED-4. Mol Cell 2004; 15: 999–1006.
Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S . Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem 2004; 73: 39–85.
Assefa Z, Van Laethem A, Garmyn M, Agostinis P . Ultraviolet radiation-induced apoptosis in keratinocytes: on the role of cytosolic factors. Biochim Biophys Acta 2005; 1755: 90–106.
Balajee AS, Bohr VA . Genomic heterogeneity of nucleotide excision repair. Gene 2000; 250: 15–30.
Prakash S, Prakash L . Nucleotide excision repair in yeast. Mutat Res 2000; 451: 13–24.
Costa RMA, Chiganças V, Galhardo R, Carvalho H, Menck CFM . The eukaryotic nucleotide excision repair pathway. Biochimie 2003; 85: 1083–1099.
Yang J, Yu Y, Hamrick HE, Duerksen-Hughes PJ . ATM, ATR, and DNA-PK: initiators of the cellular genotoxic stress responses. Carcinogenesis 2003; 24: 1571–1580.
Shiloh Y . ATM and ATR: networking cellular responses to DNA damage. Curr Opin Genet Dev 2001; 11: 71–77.
Gartner A, Milstein S, Ahmed S, Hodgkin J, Hengartner MO . A conserved checkpoint pathway mediates DNA damage--induced apoptosis and cell cycle arrest in C. elegans. Mol Cell 2000; 5: 435–443.
Schumacher B, Schertel C, Wittenburg N, Tuck S, Mitani S, Gartner A et al. C. elegans ced-13 can promote apoptosis and is induced in response to DNA damage. Cell Death Differ 2005; 12: 153–161.
Chehab NH, Malikzay A, Stavridi ES, Halazonetis TD . Phosphorylation of Ser-20 mediates stabilization of human p53 in response to DNA damage. Proc Natl Acad Sci USA 1999; 96: 13777–13782.
Matsuoka S, Huang M, Elledge SJ . Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science 1998; 282: 1893–1897.
Hirao A, Cheung A, Duncan G, Girard PM, Elia AJ, Wakeham A et al. Chk2 is a tumor suppressor that regulates apoptosis in both an ataxia telangiectasia mutated (ATM)-dependent and an ATM-independent manner. Mol Cell Biol 2002; 22: 6521–6532.
MacQueen AJ, Villeneuve AM . Nuclear reorganization and homologous chromosome pairing during meiotic prophase require C. elegans chk-2. Genes Dev 2001; 15: 1674–1687.
Liu Q, Guntuku S, Cui XS, Matsuoka S, Cortez D, Tamai K et al. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev 2000; 14: 1448–1459.
Matsuoka S, Rotman G, Ogawa A, Shiloh Y, Tamai K, Elledge SJ . Ataxia telangiectasia-mutated phosphorylates Chk2 in vivo and in vitro. Proc Natl Acad Sci USA 2000; 97: 10389–10394.
Bartek J, Falck J, Lukas J . CHK2 kinase – a busy messenger. Nat Rev Mol Cell Biol 2001; 2: 877–886.
Boulton SJ, Gartner A, Reboul J, Vaglio P, Dyson N, Hill DE et al. Combined functional genomic maps of the C. elegans DNA damage response. Science 2002; 295: 127–131.
Garcia-Muse T, Boulton SJ . Distinct modes of ATR activation after replication stress and DNA double-strand breaks in Caenorhabditis elegans. EMBO J 2005; 24: 4345–4355.
Thoma BS, Vasquez KM . Critical DNA damage recognition functions of XPC-hHR23B and XPA-RPA in nucleotide excision repair. Mol Carcinog 2003; 38: 1–13.
Park HK, Yook JS, Koo HS, Choi IS, Ahn B . The Caenorhabditis elegans XPA homolog of human XPA. Mol Cells 2002; 14: 50–55.
Bomgarden RD, Lupardus PJ, Soni DV, Yee MC, Ford JM, Cimprich KA . Opposing effects of the UV lesion repair protein XPA and UV bypass polymerase eta on ATR checkpoint signaling. EMBO J 2006; 25: 2605–2614.
de Boer J, Hoeijmakers JHJ . Nucleotide excision repair and human syndromes. Carcinogenesis 2000; 21: 453–460.
McKay BC, Becerril C, Spronck JC, Ljungman M . Ultraviolet light-induced apoptosis is associated with S-phase in primary human fibroblasts. DNA Repair (Amsterdam) 2002; 1: 811–820.
Dunkern TR, Kaina B . Cell proliferation and DNA breaks are involved in ultraviolet light-induced apoptosis in nucleotide excision repair-deficient Chinese hamster cells. Mol Biol Cell 2002; 13: 348–361.
Stout GJ, Oosten M, Acherrat FZ, Wit J, Vermeij WP, Mullenders LH et al. Selective DNA damage responses in murine Xpa(−/−), Xpc(−/−) and Csb(−/−) keratinocyte cultures. DNA Repair (Amsterdam) 2005; 4: 1337–1344.
Heyer BS, MacAuley A, Behrendtsen O, Werb Z . Hypersensitivity to DNA damage leads to increased apoptosis during early mouse development. Genes Dev 2000; 14: 2072–2084.
Weiss RS, Enoch T, Leder P . Inactivation of mouse Hus1 results in genomic instability and impaired responses to genotoxic stress. Genes Dev 2000; 14: 1886–1898.
Ahn J, Urist M, Prives C . The Chk2 protein kinase. DNA Repair (Amsterdam) 2004; 3: 1039–1047.
Brenner S . The genetics of Caenorhabditis elegans. Genetics 1974; 77: 71–94.
Gumienny TL, Lambie E, Hartwieg E, Horvitz HR, Hengartner MO . Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development 1999; 126: 1011–1022.
Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 2003; 421: 231–237.
Acknowledgements
We thank A Gartner, J Phelan, E Bogan and members of the Hengartner lab for discussions and critical comments on this paper. Some C. elegans strains were obtained from the Caenorhabditis Genetics Center, which is funded by the National Institute of Health (NIH) National Center for Research Resources (NCRR), and from the Shohei Mitani mutant collection of the National Bioresource Project in Japan. This work was supported by the Kanton of Zurich, the Swiss National Science Foundation, the Ernst Hadorn Foundation and the Josef Steiner Cancer Research Foundation. AS was supported by a MD-PhD fellowship from the Swiss National Science Foundation and Oncosuisse.
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by E Baehrecke
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Rights and permissions
About this article
Cite this article
Stergiou, L., Doukoumetzidis, K., Sendoel, A. et al. The nucleotide excision repair pathway is required for UV-C-induced apoptosis in Caenorhabditis elegans. Cell Death Differ 14, 1129–1138 (2007). https://doi.org/10.1038/sj.cdd.4402115
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.cdd.4402115
Keywords
This article is cited by
-
Mapping enzyme activity in living systems by real-time mid-infrared photothermal imaging of nitrile chameleons
Nature Methods (2024)
-
C. elegans TFIIH subunit GTF-2H5/TTDA is a non-essential transcription factor indispensable for DNA repair
Communications Biology (2021)
-
JMJD-1.2 controls multiple histone post-translational modifications in germ cells and protects the genome from replication stress
Scientific Reports (2018)
-
Dynein links engulfment and execution of apoptosis via CED-4/Apaf1 in C. elegans
Cell Death & Disease (2018)
-
Cysteine protease cathepsin B mediates radiation-induced bystander effects
Nature (2017)