Abstract
Background
How gastric cancer (GC) incidence is associated with changes in the gastric microbiome has not been firmly established. The present study therefore aims to investigate the microbial communities present within the gastric mucosa of patients with superficial gastritis (SG) or GC.
Methods
Paired tumor and paracancerous samples of the gastric mucosa were collected from 18 patients being surgically treated for GC and from 32 patients with SG being treated via gastroscopy. The gastric microbiome in these samples was then profiled via 16S rRNA sequencing, with a linear discriminant analysis effect size (LEfSe) approach used to identify and compare different bacteria, and with PICRUSt used for predictive functional analyses.
Results
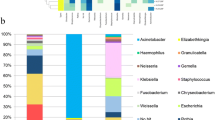
GC patients exhibited a distinct gastric microbiota profile from that observed in SG patients. These changes were evident in both tumor and paracancerous tissues from GC patients. Specifically, we found that 6 bacterial genera were specifically enriched in GC tissue samples relative to SG samples, while 18 genera were depleted in these same samples. Based on the differential abundance of these bacteria, we were able to calculate microbial dysbiosis index (MDI) values, which were significantly higher in GC patients than in SG patients. In addition, MDI values were negatively correlated with gastric Shannon index and were positively correlated with relative Helicobacter spp. abundance. Importantly, these MDI values were readily able to discriminate between GC and SG patient samples. Functional analysis suggested that GC patients were more likely to harbor a nitrosating microbial community.
Conclusions
GC patients exhibited a gastric microbiome profile distinct from that observed in SG patients, with these differences being evident in both tumor and paracancerous tissues. Differences in the relative abundance of Helicobacter spp. may be the primary driver of gastric dysbiosis in GC patients.





Similar content being viewed by others
References
Shah MA. Update on metastatic gastric and esophageal cancers. J Clin Oncol. 2015;33:1760–1769.
Correa P. Human gastric carcinogenesis: a multistep and multifactorial process–First American Cancer Society Award lecture on cancer epidemiology and prevention. Cancer Res. 1992;52:6735–6740.
Plummer M, Franceschi S, Vignat J, et al. Global burden of gastric cancer attributable to Helicobacter pylori. Int J Cancer. 2015;136:487–490.
Herrera V, Parsonnet J. Helicobacter pylori and gastric adenocarcinoma. Clin Microbiol Infect. 2009;15:971–976.
Fukase K, Kato M, Kikuchi S, et al. Effect of eradication of Helicobacter pylorion incidence of metachronous gastric carcinoma after endoscopic resection of early gastric cancer: an open-label, randomised controlled trial. Lancet. 2008;372:392–397.
Ma JL, Zhang L, Brown LM, et al. Fifteen-year effects of Helicobacter pylori, garlic, and vitamin treatments on gastric cancer incidence and mortality. J Natl Cancer Inst. 2012;104:488–492.
Lertpiriyapong K, Whary MT, Muthupalani S, et al. Gastric colonisation with a restricted commensal microbiota replicates the promotion of neoplastic lesions by diverse intestinal microbiota in the Helicobacter pylori INS-GAS mouse model of gastric carcinogenesis. Gut. 2014;63:54–63.
Lofgren JL, Whary MT, Ge Z, et al. Lack of commensal flora in Helicobacter pylori-infected INS-GAS mice reduces gastritis and delays intraepithelial neoplasia. Gastroenterology. 2011;140:210–220.
Ferreira RM, Pereira-Marques J, Pinto-Ribeiro I, et al. Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut. 2018;67:226–236.
Coker OO, Dai Z, Nie Y, et al. Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut. 2018;67:1024–1032.
Thorell K, Bengtsson-Palme J, Liu OH, et al. In vivo analysis of the viable microbiota and Helicobacter pylori transcriptome in gastric infection and early stages of carcinogenesis. Infect Immun. 2017. https://doi.org/10.1128/IAI.00031-17.
Sohn SH, Kim N, Jo HJ, et al. Analysis of gastric body microbiota by pyrosequencing: possible role of bacteria other than Helicobacter pylori in the gastric carcinogenesis. J Cancer Prev. 2017;22:115–125.
Castano-Rodriguez N, Goh KL, Fock KM, et al. Dysbiosis of the microbiome in gastric carcinogenesis. Sci Rep. 2017;7:15957.
Eun CS, Kim BK, Han DS, et al. Differences in gastric mucosal microbiota profiling in patients with chronic gastritis, intestinal metaplasia, and gastric cancer using pyrosequencing methods. Helicobacter. 2014;19:407–416.
Liu X, Shao L, Liu X et al. Alterations of gastric mucosal microbiota across different stomach microhabitats in a cohort of 276 patients with gastric cancer. EBioMedicine. 2019;40:336–348.
Dong Q, Xin Y, Wang L, et al. Characterization of gastric microbiota in twins. Curr Microbiol. 2017;74:224–229.
Dixon MF, Genta RM, Yardley JH, Correa P. Histological classification of gastritis and Helicobacter pylori infection: an agreement at last? The international workshop on the histopathology of gastritis. Helicobacter. 1997;2:S17–S24.
Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461.
Amato KR, Yeoman CJ, Kent A, et al. Habitat degradation impacts black howler monkey (Alouatta pigra) gastrointestinal microbiomes. ISME J. 2013;7:1344–1353.
Segata N, Izard J, Waldron L, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60.
Gevers D, Kugathasan S, Denson LA, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15:382–392.
Langille MG, Zaneveld J, Caporaso JG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–821.
Chen XH, Wang A, Chu AN, et al. Mucosa-associated microbiota in gastric cancer tissues compared with non-cancer tissues. Front Microbiol. 2019;10:1261.
Liu X, Shao L, Liu X, et al. Alterations of gastric mucosal microbiota across different stomach microhabitats in a cohort of 276 patients with gastric cancer. EBioMedicine. 2019;40:336–348.
Hsieh YY, Tung SY, Pan HY, et al. Increased abundance of clostridium and fusobacterium in gastric microbiota of patients with gastric cancer in Taiwan. Sci Rep. 2018;8:158.
Flemer B, Lynch DB, Brown JM, et al. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut. 2017;66:633–643.
Carding S, Verbeke K, Vipond DT, et al. Dysbiosis of the gut microbiota in disease. Microb Ecol Health Dis. 2015;26:26191.
Sears CL, Garrett WS. Microbes, microbiota, and colon cancer. Cell Host Microbe. 2014;15:317–328.
Maldonado-Contreras A, Goldfarb KC, Godoy-Vitorino F, et al. Structure of the human gastric bacterial community in relation to Helicobacter pylori status. ISME J. 2011;5:574–579.
Guo Y, Zhang Y, Gerhard M, et al. Effect of Helicobacter pylori on gastrointestinal microbiota: a population-based study in Linqu, a high-risk area of gastric cancer. Gut. 2019. https://doi.org/10.1136/gutjnl-2019-319696.
Engstrand L, Lindberg M. Helicobacter pylori and the gastric microbiota. Best Pract Res Clin Gastroenterol. 2013;27:39–45.
Gagliani N, Hu B, Huber S, et al. The fire within: microbes inflame tumors. Cell. 2014;157:776–783.
Hu YL, Pang W, Huang Y, et al. The gastric microbiome is perturbed in advanced gastric adenocarcinoma identified through shotgun metagenomics. Front Cell Infect Microbiol. 2018;8:433.
Acknowledgments
This work was supported by the Natural Science Foundation of Jiangsu Commission of Health (K2019029); Youth Projects of Jiangsu traditional Chinese Medicine Hospital (Y2018CX83); Jiangsu Province Graduate Student Practice Innovation Plan (SJCX19_0402).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10620_2020_6415_MOESM1_ESM.pdf
Supplementary Figure S1: Beta diversity analysis of the gastric microbiota. PCoA plots of the unweighted (A) and weighted (B) UniFrac distances in the full sample set in which samples were coloured by increasing age. Mantel correlations controlled with 104 permutations were used to compare distances. PCoA plots of the unweighted (C) and weighted UniFrac (D) distances in the full sample set. Samples were coloured by gender (PDF 372 kb)
10620_2020_6415_MOESM2_ESM.pdf
Supplementary Figure S2: PCoA plots of the unweighted (A) and weighted (B) UniFrac distances in gastric carcinoma. Samples were coloured by histological type. PCoA plots of the unweighted (C) and weighted (D) UniFrac distances in gastric carcinoma. Samples were coloured by tumour location. PCoA plots of the unweighted (E) and weighted (F) UniFrac distances of non-tumor tissue in gastric carcinoma. Samples were coloured by stomach anatomic sites. PCoA plots of the unweighted (G) and weighted (H) UniFrac distances in superficial gastritis. Samples were coloured by stomach anatomic sites (PDF 471 kb)
10620_2020_6415_MOESM3_ESM.pdf
Supplementary Figure S3: Different bacterial taxa in tumor and non-tumor tissues of gastric cancer patients. (A) Differential bacteria between the tumor and non-tumor tissues of gastric cancer patients by LEfSe analysis (LDA scores >3.0). Green indicates taxa enriched in tumor tissues and red indicates taxa enriched in non-tumor tissues. (B) Cladogram representation of the gastric microbiota taxa associated with tumor and non-tumor tissues of gastric cancer patients (PDF 1892 kb)
Rights and permissions
About this article
Cite this article
Wu, ZF., Zou, K., Wu, GN. et al. A Comparison of Tumor-Associated and Non-Tumor-Associated Gastric Microbiota in Gastric Cancer Patients. Dig Dis Sci 66, 1673–1682 (2021). https://doi.org/10.1007/s10620-020-06415-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10620-020-06415-y