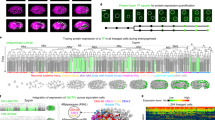
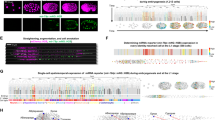
Abstract
Postembryonic development is an important process of organismal maturation after embryonic growth. Despite key progress in recent years in understanding embryonic development via fluorescence time-lapse microscopy, comparatively less live-cell imaging of postembryonic development has been done. Here we describe a protocol to image larval development in the nematode Caenorhabditis elegans. Our protocol describes the construction of fluorescent transgenic C. elegans, immobilization of worm larvae and time-lapse microscopy analysis. To improve the throughput of imaging, we developed a C. elegans triple-fluorescence imaging approach with a worm-optimized blue fluorescent protein (TagBFP), green fluorescent protein (GFP) and mCherry. This protocol has been previously applied to time-lapse imaging analysis of Q neuroblast asymmetric division, migration and apoptosis, and we show here that it can also be used to image neuritogenesis in the L1 larvae. Other applications are also possible. The protocol can be completed within 3 h and may provide insights into understanding postembryonic development.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Green, R.A. et al. Expression and imaging of fluorescent proteins in the C. elegans gonad and early embryo. Methods Cell Biol. 85, 179–218 (2008).
Sulston, J.E. & Horvitz, H.R. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56, 110–156 (1977).
Sulston, J.E., Schierenberg, E., White, J.G. & Thomson, J.N. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100, 64–119 (1983).
Ou, G. & Vale, R.D. Molecular signatures of cell migration in C. elegans Q neuroblasts. J. Cell Biol. 185, 77–85 (2009).
Ou, G., Stuurman, N., D'Ambrosio, M. & Vale, R.D. Polarized myosin produces unequal-size daughters during asymmetric cell division. Science 330, 677–680 (2010).
Singhvi, A. et al. The Arf GAP CNT-2 regulates the apoptotic fate in C. elegans asymmetric neuroblast divisions. Curr. Biol. 21, 948–954 (2011).
Li, W. et al. Autophagy genes function sequentially to promote apoptotic cell corpse degradation in the engulfing cell. J. Cell Biol. 197, 27–35 (2012).
Cheeseman, I.M. & Desai, A. A combined approach for the localization and tandem affinity purification of protein complexes from metazoans. Sci. STKE 2005, pl1 (2005).
Knobel, K.M., Jorgensen, E.M. & Bastiani, M.J. Growth cones stall and collapse during axon outgrowth in Caenorhabditis elegans. Development 126, 4489–4498 (1999).
Ou, G., Blacque, O.E., Snow, J.J., Leroux, M.R. & Scholey, J.M. Functional coordination of intraflagellar transport motors. Nature 436, 583–587 (2005).
Joshi, P.M., Riddle, M.R., Djabrayan, N.J. & Rothman, J.H. Caenorhabditis elegans as a model for stem cell biology. Dev. Dyn. 239, 1539–1554 (2010).
Pederson, T. The nucleolus. Cold Spring Harb. Perspect. Biol. 3 pii:a000638 (2011).
Goldsmith, A.D., Sarin, S., Lockery, S. & Hobert, O. Developmental control of lateralized neuron size in the nematode Caenorhabditis elegans. Neural Dev. 5, 33 (2010).
Subach, O.M. et al. Conversion of red fluorescent protein into a bright blue probe. Chem. Biol. 15, 1116–1124 (2008).
Reboul, J. et al. C. elegans ORFeome version 1.1: experimental verification of the genome annotation and resource for proteome-scale protein expression. Nat. Genet. 34, 35–41 (2003).
Reboul, J. et al. Open-reading-frame sequence tags (OSTs) support the existence of at least 17,300 genes in C. elegans. Nat. Genet. 27, 332–336 (2001).
Tursun, B., Cochella, L., Carrera, I. & Hobert, O. A toolkit and robust pipeline for the generation of fosmid-based reporter genes in C. elegans. PLoS One 4, e4625 (2009).
Hobert, O. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques 32, 728–730 (2002).
Mello, C. & Fire, A. DNA transformation. Methods Cell Biol. 48, 451–482 (1995).
Frokjaer-Jensen, C. et al. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 40, 1375–1383 (2008).
Bloom, L. & Horvitz, H.R. The Caenorhabditis elegans gene unc-76 and its human homologs define a new gene family involved in axonal outgrowth and fasciculation. Proc. Natl. Acad. Sci. USA 94, 3414–3419 (1997).
Middelkoop, T.C. et al. The thrombospondin repeat containing protein MIG-21 controls a left-right asymmetric Wnt signaling response in migrating C. elegans neuroblasts. Dev. Biol. 361, 338–348 (2012).
Chronis, N., Zimmer, M. & Bargmann, C.I. Microfluidics for in vivo imaging of neuronal and behavioral activity in Caenorhabditis elegans. Nat. Methods 4, 727–731 (2007).
Gilleland, C.L., Rohde, C.B., Zeng, F. & Yanik, M.F Microfluidic immobilization of physiologically active Caenorhabditis elegans. Nat. Protoc. 5, 1888–1902 (2010).
Stiernagle, T. Maintenance of C. elegans. in WormBook, 1–11 (ed. The C. elegans Research Community) (2006).
Cordes, S., Frank, C.A. & Garriga, G. The C. elegans MELK ortholog PIG-1 regulates cell size asymmetry and daughter cell fate in asymmetric neuroblast divisions. Development 133, 2747–2756 (2006).
Shakir, M.A., Gill, J.S. & Lundquist, E.A. Interactions of UNC-34 enabled with Rac GTPases and the NIK kinase MIG-15 in Caenorhabditis elegans axon pathfinding and neuronal migration. Genetics 172, 893–913 (2006).
Chapman, J.O., Li, H. & Lundquist, E.A. The MIG-15 NIK kinase acts cell-autonomously in neuroblast polarization and migration in C. elegans. Dev. Biol. 324, 245–257 (2008).
Honigberg, L. & Kenyon, C. Establishment of left/right asymmetry in neuroblast migration by UNC-40/DCC, UNC-73/Trio and DPY-19 proteins in C. elegans. Development 127, 4655–4668 (2000).
Wolf, F.W., Hung, M.S., Wightman, B., Way, J. & Garriga, G. vab-8 is a key regulator of posteriorly directed migrations in C. elegans and encodes a novel protein with kinesin motor similarity. Neuron 20, 655–666 (1998).
Zipkin, I.D., Kindt, R.M. & Kenyon, C.J. Role of a new Rho family member in cell migration and axon guidance in C. elegans. Cell 90, 883–894 (1997).
Baum, P.D. & Garriga, G. Neuronal migrations and axon fasciculation are disrupted in ina-1 integrin mutants. Neuron 19, 51–62 (1997).
Silhankova, M. & Korswagen, H.C. Migration of neuronal cells along the anterior-posterior body axis of C. elegans: Wnts are in control. Curr. Opin. Genet. Dev. 17, 320–325 (2007).
Kenyon, C. A gene involved in the development of the posterior body region of C. elegans. Cell 46, 477–487 (1986).
Whangbo, J. & Kenyon, C. A Wnt signaling system that specifies two patterns of cell migration in C. elegans. Mol. Cell 4, 851–858 (1999).
Pan, C.L. et al. Multiple Wnts and frizzled receptors regulate anteriorly directed cell and growth cone migrations in Caenorhabditis elegans. Dev. Cell 10, 367–377 (2006).
Kim, S., Ren, X.C., Fox, E. & Wadsworth, W.G. SDQR migrations in Caenorhabditis elegans are controlled by multiple guidance cues and changing responses to netrin UNC-6. Development 126, 3881–3890 (1999).
Acknowledgements
G.O. thanks R. Vale for his support in developing the Q cell project when G.O. was a postdoctoral fellow in his laboratory, and also thanks N. Stuurman and other Vale laboratory members for their encouragement and discussion. We thank G. Garriga, J. Teuliere, R. Korswagen, T. Middelkoop and R. Mentink for their critical comments on the manuscript. We thank T. Xu, W. Ji and X. Zhang for the microscopy setup. This work was supported by funding from the National Basic Research Program of China to W.L. and G.O. (973 Program, 2012CB966800 and 2012CB945002); the National Natural Science Foundation of China to W.L., Y.Y. and G.O. (31101002, 31100972, 31171295 and 31190063); the Natural Science Foundation of Beijing to X.W; and the Junior Thousand Talents Program of China to G.O.
Author information
Authors and Affiliations
Contributions
G.O. developed the protocol and supervised the project. Y.C., W.L., G.F., Y.Y. and X.W. performed the experiments. G.O., Y.C. and W.L. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figure 1
Sequence of ceBFP, blue fluorescent protein (TagBFP) modified by C. elegans codon optimization and intron insertion. The three artificial introns are in lower case. (PDF 429 kb)
Supplementary Figure 2
Preparation of agarose pads for mounting C. elegans in microwell dishes. (a) A Pasteur pipet was used to put one drop of 3% agarose in water on the center of the 12-mm round coverslip. (b)-(c) Another 12-mm round coverslip was placed on the top of the agarose to make a "sandwich". The agarose pad sandwich can be stored in a moist chamber (d) or can be directly opened to use (e). (PDF 558 kb)
Supplementary Table 1
Vector list for the expression of fluorescent fusion constructs in the C. elegans larva (PDF 284 kb)
Supplementary Table 2
Primer sequences to amplify fluorescence tags (PDF 326 kb)
Supplementary Table 3
Primer sequences to amplify promoters (PDF 326 kb)
Supplementary Video 1
QR cell divides. C. elegans expressing GFP-tagged CMD-1 (green) and mCherry-tagged membrane and histones in Q cells (red) were imaged in 20-second intervals over 800 seconds. Bar=5 μm. Strains in Videos 1 and 2; GOU60 casIs22 [Pegl-17::myri-mCherry + Pegl-17::mCherry-TEV-S::his-24 + Pegl-17::GFP-TEV-S::cmd-1 + rol-6(+)]. (MOV 73 kb)
Supplementary Video 2
QR.pa cell divides. C. elegans expressing GFP-tagged CMD-1 (green) and mCherry-tagged membrane and histone in Q cells (red) were imaged in 30-second intervals over 360 seconds. Bar=5 μm. (MOV 24 kb)
Supplementary Video 3
QR.ap cell migrates. C. elegans expressing mCherry-tagged membrane and histones in Q cells (red) were imaged in 1-minute intervals over 60 minutes. Bar=5 μm. Strains; RDV55 rdvIs1[Pegl-17::myri-mCherry + Pegl-17::mCherry-TEV-S::his-24 + Pegl-17::mig-10::YFP + rol-6(+)]. (MOV 266 kb)
Supplementary Video 4
QR. ap/AQR dendrite grows. C. elegans expressing mCherry-tagged membrane in Q cells (red) were imaged in 1 minute intervals over 53 minutes. Bar=5 μm. Strain; GOU286 casIs38[Pegl-17::myri-mCherry + Pegl-17::mCherry-TEV-S::his-24 + Pegl-17::vab-8::GFP + unc-76(+)]. (MOV 181 kb)
Supplementary Video 5
Actin and EPG-5 co-recruit on QR.aa corpse. C. elegans expressing BFP-tagged actin (blue) and GFP-tagged EPG-5 (green) in the hyp7 cell and mCherry in Q cells (red) were imaged in 1 minute intervals over 18 minutes. Bar=5 μm. Strain; GOU502 casEx489 [Pegl-17::myri-mCherry + Pegl-17::mCherry-TEV-S::his-24 + PY37A1B.5::BFP-TEV-S::act-1 + Pced-1::epg-5::GFP + unc-76(+)]. (MOV 27 kb)
Rights and permissions
About this article
Cite this article
Chai, Y., Li, W., Feng, G. et al. Live imaging of cellular dynamics during Caenorhabditis elegans postembryonic development. Nat Protoc 7, 2090–2102 (2012). https://doi.org/10.1038/nprot.2012.128
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2012.128
This article is cited by
-
Dual-expression system for blue fluorescent protein optimization
Scientific Reports (2022)
-
Wnt signaling polarizes cortical actin polymerization to increase daughter cell asymmetry
Cell Discovery (2022)
-
Live-cell confocal microscopy and quantitative 4D image analysis of anchor-cell invasion through the basement membrane in Caenorhabditis elegans
Nature Protocols (2017)
-
Long-term time-lapse live imaging reveals extensive cell migration during annelid regeneration
BMC Developmental Biology (2016)
-
Long-term time-lapse microscopy of C. elegans post-embryonic development
Nature Communications (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.