Abstract
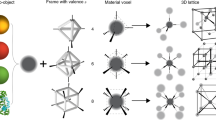
Three-dimensional mesoscale clusters that are formed from nanoparticles spatially arranged in pre-determined positions can be thought of as mesoscale analogues of molecules. These nanoparticle architectures could offer tailored properties due to collective effects, but developing a general platform for fabricating such clusters is a significant challenge. Here, we report a strategy for assembling three-dimensional nanoparticle clusters that uses a molecular frame designed with encoded vertices for particle placement. The frame is a DNA origami octahedron and can be used to fabricate clusters with various symmetries and particle compositions. Cryo-electron microscopy is used to uncover the structure of the DNA frame and to reveal that the nanoparticles are spatially coordinated in the prescribed manner. We show that the DNA frame and one set of nanoparticles can be used to create nanoclusters with different chiroptical activities. We also show that the octahedra can serve as programmable interparticle linkers, allowing one- and two-dimensional arrays to be assembled with designed particle arrangements.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Halverson, J. D. & Tkachenko, A. V. DNA-programmed mesoscopic architecture. Phys. Rev. E 87, 062310 (2013).
Zeravcic, Z. & Brenner, M. P. Self-replicating colloidal clusters. Proc. Natl Acad. Sci. USA 111, 1748–1753 (2013).
Maye, M. M., Gang, O. & Cotlet, M. Photoluminescence enhancement in CdSe/ZnS-DNA linked-Au nanoparticle heterodimers probed by single molecule spectroscopy. Chem. Commun. 46, 6111–6113 (2010).
Yan, W. et al. Self-assembly of chiral nanoparticle pyramids with strong R/S optical activity. J. Am. Chem. Soc. 134, 15114–15121 (2012).
Fan, J. A. et al. Plasmonic mode engineering with templated self-assembled nanoclusters. Nano Lett. 12, 5318–5324 (2012).
Hiroi, K., Komatsu, K. & Sato, T. Superspin glass originating from dipolar interaction with controlled interparticle distance among gamma-Fe2O3 nanoparticles with silica shells. Phys. Rev. B 83, 224423 (2011).
Govorov, A. O., Fan, Z., Hernandez, P., Slocik, J. M. & Naik, R. R. Theory of circular dichroism of nanomaterials comprising chiral molecules and nanocrystals: plasmon enhancement, dipole interactions, and dielectric effects. Nano Lett. 10, 1374–1382 (2010).
Kuzyk, A. et al. DNA-based self-assembly of chiral plasmonic nanostructures with tailored optical response. Nature 483, 311–314 (2012).
Barrow, S. J., Wei, X., Baldauf, J. S., Funston, A. M. & Mulvaney, P. The surface plasmon modes of self-assembled gold nanocrystals. Nature Commun. 3, 1–9 (2012).
Ruzicka, B. et al. Observation of empty liquids and equilibrium gels in a colloidal clay. Nature Mater. 10, 56–60 (2011).
Wang, Y. et al. Colloids with valence and specific directional bonding. Nature 491, 51–56 (2012).
Chen, Q. et al. Supracolloidal reaction kinetics of janus spheres. Science 331, 199–202 (2011).
Mastroianni, A. J., Claridge, S. A. & Alivisatos, A. P. Pyramidal and chiral groupings of gold nanocrystals assembled using DNA scaffolds. J. Am. Chem. Soc. 131, 8455–8459 (2009).
Klinkova, A., Thérien-Aubin, H., Choueiri, R. M., Rubinstein, M. & Kumacheva, E. Colloidal analogs of molecular chain stoppers. Proc. Natl Acad. Sci. USA 110, 18775–18779 (2013).
Kim, J-W., Kim, J-H. & Deaton, R. DNA-linked nanoparticle building blocks for programmable matter. Angew. Chem. Int. Ed. 50, 9185–9190 (2011).
Alivisatos, A. P. et al. Organization of ‘nanocrystal molecules’ using DNA. Nature 382, 609–611 (1996).
Maye, M. M., Nykypanchuk, D., Cuisinier, M., van der Lelie, D. & Gang, O. Stepwise surface encoding for high-throughput assembly of nanoclusters. Nature Mater. 8, 388–391 (2009).
Pal, S., Deng, Z., Ding, B., Yan, H. & Liu, Y. DNA-origami-directed self-assembly of discrete silver-nanoparticle architectures. Angew. Chem. Int. Ed. 49, 2700–2704 (2010).
Lee, J., Hernandez, P., Govorov, A. O. & Kotov, N. A. Exciton–plasmon interactions in molecular spring assemblies of nanowires and wavelength-based protein detection. Nature Mater. 6, 291–295 (2007).
Sun, D. et al. Heterogeneous nanoclusters assembled by PNA-templated double-stranded DNA. Nanoscale 4, 6722–6725 (2012).
Zhang, Y., Lu, F., Yager, K. G., van der Lelie, D. & Gang, O. A general strategy for the DNA-mediated self-assembly of functional nanoparticles into heterogeneous systems. Nature Nanotech. 8, 865–872 (2013).
Zhang, C. et al. A general approach to DNA-programmable atom equivalents. Nature Mater. 12, 741–746 (2013).
Seeman, N. C. DNA in a material world. Nature 421, 427–431 (2003).
Shih, W. M., Quispe, J. D. & Joyce, G. F. A 1.7-kilobase single-stranded DNA that folds into a nanoscale octahedron. Nature 427, 618–621 (2004).
Zheng, J. et al. From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal. Nature 461, 74–77 (2009).
Nykypanchuk, D., Maye, M. M., van der Lelie, D. & Gang, O. DNA-guided crystallization of colloidal nanoparticles. Nature 451, 549–552 (2008).
Park, S. Y. et al. DNA-programmable nanoparticle crystallization. Nature 451, 553–556 (2008).
Rothemund, P. W. K. Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006).
Dietz, H., Douglas, S. M. & Shih, W. M. Folding DNA into twisted and curved nanoscale shapes. Science 325, 725–730 (2009).
Zhang, C. et al. DNA nanocages swallow gold nanoparticles (AuNPs) to form AuNP@DNA cage core–shell structures. ACS Nano 8, 1130–1135 (2014).
Sharma, J. et al. Control of self-assembly of DNA tubules through integration of gold nanoparticles. Science 323, 112–116 (2009).
Andersen, E. S. et al. Self-assembly of a nanoscale DNA box with a controllable lid. Nature 459, 73–76 (2009).
He, Y. et al. Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra. Nature 452, 198–201 (2008).
Bai, X., Martin, T. G., Scheres, S. H. W. & Dietz, H. Cryo-EM structure of a 3D DNA-origami object. Proc. Natl Acad. Sci. USA 149, 20012–20017 (2012).
Midgley, P. A. & Dunin-Borkowski, R. E. Electron tomography and holography in materials science. Nature Mater. 8, 271–280 (2009).
Kourkoutis, L. F., Plitzko, J. M. & Baumeister, W. Electron microscopy of biological materials at the nanometer scale. Annu. Rev. Mater. Res. 42, 33–58 (2012).
De-Rosier, D. & Klug, A. Reconstruction of three dimensional structures from electron micrographs. Nature 217, 130–134 (1968).
Douglas, S. M. et al. Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res. 37, 5001–5006 (2009).
Mathieu, F. et al. Six-helix bundles designed from DNA. Nano Lett. 5, 661–665 (2005).
Douglas, S. M. et al. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 459, 414–418 (2009).
Douglas, S. M., Chou, J. J. & Shih, W. M. DNA-nanotube-induced alignment of membrane proteins for NMR structure determination. Proc. Natl Acad. Sci. USA 104, 6644–6648 (2006).
Hill, H. D., Millstone, J. E., Banholzer, M. J. & Mirkin, C. A. The role radius of curvature plays in thiolated oligonucleotide loading on gold nanoparticles. ACS Nano 3, 418–424 (2009).
King, R. B. Nonhanded chirality in octahedral metal complexes. Chirality 13, 465–473 (2001).
Chi, C., Vargas-Lara, F., Tkachenko, A. V., Starr, F. W. & Gang, O. Internal structure of nanoparticle dimers linked by DNA. ACS Nano 6, 6793–7802 (2012).
Kaya, H. Scattering from cylinders with globular end-caps. J. Appl. Crystallogr. 37, 223–230 (2004).
Acknowledgements
The authors thank J. Li for help with tomography analysis and D. Chen for assistance with schematic drawing. Research carried out at the Centre for Functional Nanomaterials, Brookhaven National Laboratory, was supported by the US Department of Energy, Office of Basic Energy Sciences (contract no. DE-SC0012704). H.L. was supported by a National Institutes of Health R01 grant (AG029979) and W.M.S. was supported by a National Science Foundation Expeditions grant (1317694) and Designing Materials to Revolutionize and Engineer Our Future grant (1435964).
Author information
Authors and Affiliations
Contributions
Y.T. and O.G. conceived and designed the experiments. Y.T. performed the experiments. W.M.S. and Y.K. contributed to the octahedral design. Y.T., T.W., W.L. and O.G. analysed the data. T.W. and H.L. contributed to the cryo-EM measurement and reconstruction. H.X. contributed to the tomography analysis. Y.T. and O.G. wrote the paper. O.G. supervised the project. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary Information (PDF 3194 kb)
Rights and permissions
About this article
Cite this article
Tian, Y., Wang, T., Liu, W. et al. Prescribed nanoparticle cluster architectures and low-dimensional arrays built using octahedral DNA origami frames. Nature Nanotech 10, 637–644 (2015). https://doi.org/10.1038/nnano.2015.105
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nnano.2015.105
This article is cited by
-
A Review of Fabrication of DNA Origami Plasmonic Structures for the Development of Surface-Enhanced Raman Scattering (SERS) Platforms
Plasmonics (2023)
-
Chiral inorganic nanomaterials: Harnessing chirality-dependent interactions with living entities for biomedical applications
Nano Research (2023)
-
Self-assembled inorganic chiral superstructures
Nature Reviews Chemistry (2022)
-
Three-dimensional electron ptychography of organic–inorganic hybrid nanostructures
Nature Communications (2022)
-
Cryo-EM for nanomaterials: Progress and perspective
Science China Materials (2022)