Abstract
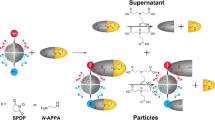
In a physiological environment, nanoparticles selectively absorb proteins to form ‘nanoparticle–protein coronas’1,2,3,4,5, a process governed by molecular interactions between chemical groups on the nanoparticle surfaces and the amino-acid residues of the proteins6,7,8. Here, we propose a biological surface adsorption index to characterize these interactions by quantifying the competitive adsorption of a set of small molecule probes onto the nanoparticles. The adsorption properties of nanomaterials are assumed to be governed by Coulomb forces, London dispersion, hydrogen-bond acidity and basicity, polarizability and lone-pair electrons. Adsorption coefficients of the probe compounds were measured and used to create a set of nanodescriptors representing the contributions and relative strengths of each molecular interaction. The method successfully predicted the adsorption of various small molecules onto carbon nanotubes, and the nanodescriptors were also measured for 12 other nanomaterials. The biological surface adsorption index nanodescriptors can be used to develop pharmacokinetic and safety assessment models for nanomaterials.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Lynch, I., Dawson, K. A. & Linse, S. Detecting cryptic epitopes created by nanoparticles. Sci. STKE 327, 14 (2006).
Cedervall, T. et al. Understanding the nanoparticle–protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles. Proc. Natl Acad. Sci. USA 104, 2050–2055 (2007).
Lynch, I. & Dawson, K. A. Protein–nanoparticle interactions. Nano Today 3, 40–47 (2008).
Lundqvist, M. et al. Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts. Proc. Natl Acad. Sci. USA 105, 14265–14270 (2008).
Hellstrand, E. et al. Complete high-density lipoproteins in nanoparticle corona. FEBS J. 276, 3372–3381 (2009).
Nel, A. E. et al. Understanding biophysicochemical interactions at the nano–bio interface. Nature Mater. 8, 543–557 (2009).
Puzyn, T., Leszczynska, D. & Leszczynski, J. Toward the development of ‘Nano-QSARs’: advances and challenges. Small 5, 2494–2509 (2009).
Aillon, K. L., Xie, Y., El-Gendy, N., Berkland, C. J. & Forrest, M. L. Effects of nanomaterial physicochemical properties on in vivo toxicity. Adv. Drug Deliv. Rev. 61, 457–466 (2009).
Yang, K. & Xing, B. Adsorption of organic compounds by carbon nanomaterials in aqueous phase: Polanyi theory and its application. Chem. Rev. doi: 10.1021/cr100059s (2010).
Aggarwal, P., Hall, J. B., McLeland, C. B., Dobrovolskaia, M. A. & McNeil, S. E. Nanoparticle interaction with plasma proteins as it relates to particle biodistribution, biocompatibility and therapeutic efficacy. Adv. Drug Deliv. Rev. 61, 428–437 (2009).
Patil, S., Sandberg, A., Heckert, E., Self, W. & Seal, S. Protein adsorption and cellular uptake of cerium oxide nanoparticles as a function of zeta potential. Biomaterials 28, 4600–4607 (2007).
Simon, P. & Joner, E. Conceivable interactions of biopersistent nanoparticles with food matrix and living systems following from their physicochemical properties. J. Food Nutr. Res. 47, 51–59 (2008).
Stone, A. J. (ed.) The Theory of Intermolecular Forces (Oxford Univ. Press, 1996).
Karelson, M. (ed.) Molecular Descriptors in QSAR/QSPR (John Wiley & Sons, 2000).
Abraham, M. H. Scales of solute hydrogen-bonding: their construction and application to physicochemical and biochemical processes. Chem. Soc. Rev. 22, 73–83 (1993).
Hansch, C. & Fujita, T. A method for the correlation of biological activity and chemical structure. J. Am. Chem. Soc. 86, 1616–1626 (1964).
Hansch, C. Quantitative structure—activity relationships and the unnamed science. Acc. Chem. Res. 26, 147–153 (1993).
Giaginis, C. & Tsantili-Kakoulidou, A. Alternative measures of lipophilicity: from octanol–water partitioning to IAM retention. J. Pharm. Sci. 97, 2984–3004 (2008).
Chen, W., Duan, L. & Zhu, D. Adsorption of polar and nonpolar organic chemicals to carbon nanotubes. Environ. Sci. Technol. 41, 8295–8300 (2007).
Arkin, M. R. et al. Rates of DNA-mediated electron transfer between metallointercalators. Science 273, 475–480 (1996).
Gramatica, P. Principles of QSAR models validation: internal and external. QSAR Comb. Sci. 26, 694–701 (2007).
Dearden, J. C., Cronin, M. T. D. & Kaiser, K. L. E. How not to develop a quantitative structure–activity or structure–property relationship (QSAR/QSPR). SAR QSAR Environ. Res. 20, 241–266 (2009).
Gramatica, P., Giani, E. & Papa, E. Statistical external validation and consensus modeling: a QSPR case study for Koc prediction. J. Mol. Graph. Model. 25, 755–766 (2007).
Duong, D. D. (ed.) Adsorption Analysis: Equilibria and Kinetics (Imperial College Press, 1998).
Yang, K., Wang, X., Zhu, L. & Xing, B. Competitive sorption of pyrene, phenanthrene and naphthalene on multiwalled carbon nanotubes. Environ. Sci. Technol. 40, 5804–5810 (2006).
Zhang, S., Shado, T., Bekaroglu, S. S. & Karanfil, T. The impacts of aggregation and surface chemistry of carbon nanotubes on the adsorption of synthetic organic compounds. Environ. Sci. Technol. 43, 5719–5725 (2009).
Johnson, R. R., Johnson, A. T. & Klein, M. L. The nature of DNA-base—carbon-nanotube interactions. Small 6, 31–34 (2010).
Xia, X. R., Baynes, R. E., Monteiro-Riviere, N. A. & Riviere, J. E. A system coefficient approach for quantitative assessment of the solvent effects on membrane absorption from chemical mixtures. SAR QSAR Environ. Res. 18, 579–593 (2007).
Lee, H. A., Leavens, T. L., Mason, S. E., Monteiro-Riviere, N. A. & Riviere, J. E. Comparison of quantum dot biodistribution with a blood-flow-limited physiologically based pharmacokinetic model. Nano Lett. 9, 794–799 (2009).
Zhang, L. W. & Monteiro-Riviere, N. A. Mechanisms of quantum dot nanoparticle cellular uptake. Toxicol. Sci. 110, 138–155 (2009).
Xia, X. R. et al. A novel in vitro technique for studying percutaneous permeation with a membrane-coated fiber and gas chromatography/mass spectrometry Part I. Performances of the technique and determination of the permeation rates and partition coefficients of chemical mixtures. Pharm. Res. 20, 272–279 (2003).
Acknowledgements
This research was supported by the USEPA STAR grant no. R833328 and the United States Air Force Office of Scientific Research (USAFOSR) grant no. FA9550-08-1-0182. The authors thank J. Brooks for help in QSAR validation techniques.
Author information
Authors and Affiliations
Contributions
X.-R.X. conceived, conducted and designed the experiments and analysed the resulting data. N.A.M.-R. initiated the application of this technique to nanomaterial characterization and provided biological context. J.E.R. conceived the BSAI concept and contributed to data analysis and interpretation. All three authors were involved in writing and revising the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary information (PDF 917 kb)
Rights and permissions
About this article
Cite this article
Xia, XR., Monteiro-Riviere, N. & Riviere, J. An index for characterization of nanomaterials in biological systems. Nature Nanotech 5, 671–675 (2010). https://doi.org/10.1038/nnano.2010.164
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nnano.2010.164
This article is cited by
-
Basic and advanced spectrometric methods for complete nanoparticles characterization in bio/eco systems: current status and future prospects
Analytical and Bioanalytical Chemistry (2023)
-
Exploring structural requirements of simple benzene derivatives for adsorption on carbon nanotubes: CoMFA, GRIND, and HQSAR
Structural Chemistry (2023)
-
Predicting adsorption of organic compounds onto graphene and black phosphorus by molecular dynamics and machine learning
Environmental Science and Pollution Research (2023)
-
QSPR models for predicting the adsorption capacity for microplastics of polyethylene, polypropylene and polystyrene
Scientific Reports (2020)
-
Conformational flexibility of fatty acid-free bovine serum albumin proteins enables superior antifouling coatings
Communications Materials (2020)