Abstract
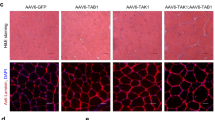
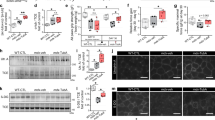
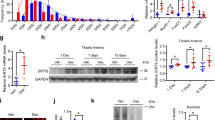
During physical exercise, increases in motor neuron activity stimulate the expression of muscle-specific genes through the myocyte enhancer factor 2 (MEF2) family of transcription factors. Elevations in intracellular calcium increase MEF2 activity via the phosphorylation-dependent inactivation of class II histone deacetylases (HDACs). In studies to determine the role of the cAMP responsive element binding protein (CREB) in skeletal muscle, we found that mice expressing a dominant-negative CREB transgene (M-ACREB mice) exhibited a dystrophic phenotype along with reduced MEF2 activity. Class II HDAC phosphorylation was decreased in M-ACREB myofibers due to a reduction in amounts of Snf1lk (encoding salt inducible kinase, SIK1), a CREB target gene that functions as a class II HDAC kinase. Inhibiting class II HDAC activity either by viral expression of Snf1lk or by the administration of a small molecule antagonist improved the dystrophic phenotype in M-ACREB mice, pointing to an important role for the SIK1-HDAC pathway in regulating muscle function.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Wu, H. et al. Activation of MEF2 by muscle activity is mediated through a calcineurin-dependent pathway. EMBO J. 20, 6414–6423 (2001).
Bassel-Duby, R. & Olson, E.N. Signaling pathways in skeletal muscle remodeling. Annu. Rev. Biochem. 75, 19–37 (2006).
Vega, R.B. et al. Protein kinases C and D mediate agonist-dependent cardiac hypertrophy through nuclear export of histone deacetylase 5. Mol. Cell. Biol. 24, 8374–8385 (2004).
Chang, S., Bezprozvannaya, S., Li, S. & Olson, E.N. An expression screen reveals modulators of class II histone deacetylase phosphorylation. Proc. Natl. Acad. Sci. USA 102, 8120–8125 (2005).
Mayr, B. & Montminy, M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell Biol. 2, 599–609 (2001).
West, A.E. et al. Calcium regulation of neuronal gene expression. Proc. Natl. Acad. Sci. USA 98, 11024–11031 (2001).
Wu, Z. et al. Transducer of regulated CREB-binding proteins (TORCs) induce PGC-1á transcription and mitochondrial biogenesis in muscle cells. Proc. Natl. Acad. Sci. USA 103, 14379–14384 (2006).
Handschin, C., Rhee, J., Lin, J., Tarr, P.T. & Spiegelman, B.M. An autoregulatory loop controls peroxisome proliferator-activated receptor gamma coactivator 1α expression in muscle. Proc. Natl. Acad. Sci. USA 100, 7111–7116 (2003).
Chen, A.E., Ginty, D.D. & Fan, C.M. Protein kinase A signalling via CREB controls myogenesis induced by Wnt proteins. Nature 433, 317–322 (2005).
Bleckmann, S.C. et al. Activating transcription factor 1 and CREB are important for cell survival during early mouse development. Mol. Cell. Biol. 22, 1919–1925 (2002).
Ahn, S. et al. A dominant-negative inhibitor of CREB reveals that it is a general mediator of stimulus-dependent transcription of c-fos. Mol. Cell. Biol. 18, 967–977 (1998).
De Cesare, D. & Sassone-Corsi, P. Transcriptional regulation by cyclic AMP-responsive factors. Prog. Nucleic Acid Res. Mol. Biol. 64, 343–369 (2000).
Lonze, B.E. & Ginty, D.D. Function and regulation of CREB family transcription factors in the nervous system. Neuron 35, 605–623 (2002).
Hummler, E. et al. Targeted mutation of the CREB gene: compensation within the CREB/ATF family of transcription factors. Proc. Natl. Acad. Sci. USA 91, 5647–5651 (1994).
Shaywitz, A.J. & Greenberg, M.E. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu. Rev. Biochem. 68, 821–861 (1999).
Brennan, K.J. & Hardeman, E.C. Quantitative analysis of the human α-skeletal actin gene in transgenic mice. J. Biol. Chem. 268, 719–725 (1993).
Garry, D.J. et al. Myogenic stem cell function is impaired in mice lacking the forkhead/winged helix protein MNF. Proc. Natl. Acad. Sci. USA 97, 5416–5421 (2000).
Watchko, J.F., O'Day, T.L. & Hoffman, E.P. Functional characteristics of dystrophic skeletal muscle: insights from animal models. J. Appl. Physiol. 93, 407–417 (2002).
Li, H. & Capetanaki, Y. Regulation of the mouse desmin gene: transactivated by MyoD, myogenin, MRF4 and Myf5. Nucleic Acids Res. 21, 335–343 (1993).
Milner, D.J., Weitzer, G., Tran, D., Bradley, A. & Capetanaki, Y. Disruption of muscle architecture and myocardial degeneration in mice lacking desmin. J. Cell Biol. 134, 1255–1270 (1996).
Li, Z. et al. Cardiovascular lesions and skeletal myopathy in mice lacking desmin. Dev. Biol. 175, 362–366 (1996).
Zhang, X. et al. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc. Natl. Acad. Sci. USA 102, 4459–4464 (2005).
Molkentin, J.D. & Olson, E.N. Combinatorial control of muscle development by basic helix-loop-helix and MADS-box transcription factors. Proc. Natl. Acad. Sci. USA 93, 9366–9373 (1996).
McKinsey, T.A., Zhang, C.L., Lu, J. & Olson, E.N. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature 408, 106–111 (2000).
Zhang, C.L. et al. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell 110, 479–488 (2002).
Screaton, R.A. et al. The CREB coactivator TORC2 functions as a calcium- and cAMP-sensitive coincidence detector. Cell 119, 61–74 (2004).
van der Linden, A.M., Nolan, K.M. & Sengupta, P. KIN-29 SIK regulates chemoreceptor gene expression via an MEF2 transcription factor and a class II HDAC. EMBO J. 26, 358–370 (2006).
Mikkelsen, U.R., Gissel, H., Fredsted, A. & Clausen, T. Excitation-induced cell damage and β2-adrenoceptor agonist stimulated force recovery in rat skeletal muscle. Am. J. Physiol. Regul. Integr. Comp. Physiol. 290, R265–R272 (2006).
Talanian, J.L. et al. Adrenergic regulation of HSL serine phosphorylation and activity in human skeletal muscle during the onset of exercise. Am. J. Physiol. Regul. Integr. Comp. Physiol. 291, R1094–R1099 (2006).
Katoh, Y. et al. Silencing the constitutive active transcription factor CREB by the LKB1-SIK signaling cascade. FEBS J. 273, 2730–2748 (2006).
Kao, H.Y. et al. Mechanism for nucleocytoplasmic shuttling of histone deacetylase 7. J. Biol. Chem. 276, 47496–47507 (2001).
Miska, E.A. et al. Differential localization of HDAC4 orchestrates muscle differentiation. Nucleic Acids Res. 29, 3439–3447 (2001).
Takemori, H., Katoh, Y., Horike, N., Doi, J. & Okamoto, M. ACTH-induced nucleocytoplasmic translocation of salt-inducible kinase. Implication in the protein kinase A-activated gene transcription in mouse adrenocortical tumor cells. J. Biol. Chem. 277, 42334–42343 (2002).
Belfield, J.L., Whittaker, C., Cader, M.Z. & Chawla, S. Differential effects of Ca2+ and cAMP on transcription mediated by MEF2D and cAMP-response element-binding protein in hippocampal neurons. J. Biol. Chem. 281, 27724–27732 (2006).
Koo, S.H. et al. The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature 437, 1109–1111 (2005).
Fentzke, R.C., Korcarz, C.E., Lang, R.M., Lin, H. & Leiden, J.M. Dilated cardiomyopathy in transgenic mice expressing a dominant- negative CREB transcription factor in the heart. J. Clin. Invest. 101, 2415–2426 (1998).
Ruiz, J.C., Conlon, F.L. & Robertson, E.J. Identification of novel protein kinases expressed in the myocardium of the developing mouse heart. Mech. Dev. 48, 153–164 (1994).
Dubowitz, V. Histological and histochemical stains and reactions. in Muscle Biopsy. A Practical Approach (ed. Dubowitz, V.) 19–40 (Bailliere Tindall, London, 1985).
Koo, S.H. et al. PGC-1 promotes insulin resistance in liver through PPAR-α-dependent induction of TRB-3. Nat. Med. 10, 530–534 (2004).
Banaszynski, L.A., Chen, L.C., Maynard-Smith, L.A., Ooi, A.G. & Wandless, T.J. A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell 126, 995–1004 (2006).
Rabinowitz, J.E. et al. Cross-packaging of a single adeno-associated virus (AAV) type 2 vector genome into multiple AAV serotypes enables transduction with broad specificity. J. Virol. 76, 791–801 (2002).
Acknowledgements
We thank the members of the Montminy lab for helpful discussions, N. Huff for technical assistance with the histology, M. Scadeng for X-ray analysis, and X. Zhang and L. Ouyang for help with microarray experiments. We thank E. Olson (University of Texas Southwestern Medical Center) for Mef2-luciferase and Mef2c expression constructs, T. McKinsey (Myogen, Inc.) for antibodies to pHDAC, M. Okamoto (Osaka Medical School, Japan) for antibodies to SIK1, and T.P. Yao (Duke University) for recombinant HDAC4. We also thank M. Weitzman (the Salk Institute) for AAV reagents. This work was supported by grants from the National Institutes of Health (National Institute of General Medical Sciences and National Institute of Diabetes and Digestive and Kidney Diseases) and the Kieckhefer Foundation.
Author information
Authors and Affiliations
Contributions
R.B. performed animal and cell culture experiments. N.G. performed histological analyses and aided in biochemical studies. L.B. and T.W. provided expertise on the Shld-1 expression system. H.T. provided expertise and reagents for SIK1 analysis. G.D.S. performed histochemical analyses and provided expertise on muscle pathology. Experimental design, data analysis and manuscript preparation were carried out by R.B. and M.M.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
M-ACREB transgenic animals develop progressive muscular dystrophy. (PDF 155 kb)
Supplementary Fig. 2
Absence of fibrosis, changes in fiber-type distribution and metabolic storage products in M-ACREB transgenic mice. (PDF 289 kb)
Supplementary Fig. 3
MEF2 target gene expression is reduced in skeletal muscles of M-ACREB mice. (PDF 61 kb)
Supplementary Fig. 4
SIK1 protein is induced by cAMP and reduced in M-ACREB tissue. (PDF 133 kb)
Supplementary Fig. 5
SIK1 phosphorylates class II HDACs. (PDF 137 kb)
Supplementary Fig. 6
RNAi mediated knockdown of SIK1 induces necrosis of C2C12 cells. (PDF 153 kb)
Supplementary Fig. 7
AAV-mediated delivery of SIK1 rescues the dystrophic phenotype in MACREB transgenic muscle. (PDF 174 kb)
Rights and permissions
About this article
Cite this article
Berdeaux, R., Goebel, N., Banaszynski, L. et al. SIK1 is a class II HDAC kinase that promotes survival of skeletal myocytes. Nat Med 13, 597–603 (2007). https://doi.org/10.1038/nm1573
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm1573
This article is cited by
-
Designing of potent anti-diabetic molecules by targeting SIK2 using computational approaches
Molecular Diversity (2023)
-
Salt-inducible kinase 1-CREB-regulated transcription coactivator 1 signalling in the paraventricular nucleus of the hypothalamus plays a role in depression by regulating the hypothalamic–pituitary–adrenal axis
Molecular Psychiatry (2022)
-
Kinase signalling in excitatory neurons regulates sleep quantity and depth
Nature (2022)
-
Seasonal regulation of singing-driven gene expression associated with song plasticity in the canary, an open-ended vocal learner
Molecular Brain (2021)
-
Salt inducible kinases 2 and 3 are required for thymic T cell development
Scientific Reports (2021)