Abstract
Pluripotent stem cells (PSCs) are capable of dynamic interconversion between distinct substates; however, the regulatory circuits specifying these states and enabling transitions between them are not well understood. Here we set out to characterize transcriptional heterogeneity in mouse PSCs by single-cell expression profiling under different chemical and genetic perturbations. Signalling factors and developmental regulators show highly variable expression, with expression states for some variable genes heritable through multiple cell divisions. Expression variability and population heterogeneity can be influenced by perturbation of signalling pathways and chromatin regulators. Notably, either removal of mature microRNAs or pharmacological blockage of signalling pathways drives PSCs into a low-noise ground state characterized by a reconfigured pluripotency network, enhanced self-renewal and a distinct chromatin state, an effect mediated by opposing microRNA families acting on the Myc/Lin28/let-7 axis. These data provide insight into the nature of transcriptional heterogeneity in PSCs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
21 January 2015
An Erratum to this paper has been published: https://doi.org/10.1038/nature14150
References
Boyer, L. A. et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947–956 (2005)
Loh, Y.-H. et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nature Genet. 38, 431–440 (2006)
Loh, Y. H. et al. Genomic approaches to deconstruct pluripotency. Annu. Rev. Genomics Hum. Genet. 12, 165–185 (2011)
MacArthur, B. D., Ma'ayan, A. & Lemischka, I. R. Systems biology of stem cell fate and cellular reprogramming. Nature Rev. Mol. Cell Biol. 10, 672–681 (2009)
Young, R. A. Control of the embryonic stem cell state. Cell 144, 940–954 (2011)
Chambers, I. et al. Nanog safeguards pluripotency and mediates germline development. Nature 450, 1230–1234 (2007)
Hayashi, K., Lopes, S. M. C. S., Tang, F. & Surani, M. A. Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell 3, 391–401 (2008)
Hong, S.-H. et al. Cell fate potential of human pluripotent stem cells is encoded by histone modifications. Cell Stem Cell 9, 24–36 (2011)
Kalmar, T. et al. Regulated fluctuations in Nanog expression mediate cell fate decisions in embryonic stem cells. PLoS Biol. 7, e1000149 (2009)
Karwacki-Neisius, V. et al. Reduced Oct4 expression directs a robust pluripotent state with distinct signaling activity and increased enhancer occupancy by Oct4 and Nanog. Cell Stem Cell 12, 531–545 (2013)
MacArthur, B. D. et al. Nanog-dependent feedback loops regulate murine embryonic stem cell heterogeneity. Nature Cell Biol. 14, 1139–1147 (2012)
Reynolds, N. et al. NuRD suppresses pluripotency gene expression to promote transcriptional heterogeneity and lineage commitment. Cell Stem Cell 10, 583–594 (2012)
Ying, Q.-L. et al. The ground state of embryonic stem cell self-renewal. Nature 453, 519–523 (2008)
Arias, A. M. & Brickman, J. M. Gene expression heterogeneities in embryonic stem cell populations: Origin and function. Curr. Opin. Cell Biol. 23, 1–7 (2011)
Cherry, A. & Daley, G. Q. Another horse in the meta-stable state of pluripotency. Cell Stem Cell 7, 641–642 (2010)
Graf, T. & Stadtfeld, M. Heterogeneity of embryonic and adult stem cells. Cell Stem Cell 3, 480–483 (2008)
Halley, J. D. et al. Self-organizing circuitry and emergent computation in mouse embryonic stem cells. Stem Cell Res. 8, 324–333 (2012)
Loh, K. M. & Lim, B. A precarious balance: pluripotency factors as lineage specifiers. Cell Stem Cell 8, 363–369 (2011)
MacArthur, B. D. & Lemischka, I. R. Statistical mechanics of pluripotency. Cell 154, 484–489 (2013)
Silva, J. & Smith, A. Capturing pluripotency. Cell 132, 532–536 (2008)
Thomson, M. et al. Pluripotency factors in embryonic stem cells regulate differentiation into germ layers. Cell 145, 875–889 (2011)
Cahan, P. & Daley, G. Q. Origins and implications of pluripotent stem cell variability and heterogeneity. Nature Rev. Mol. Cell Biol. 14, 357–368 (2013)
Huang, S. Non-genetic heterogeneity of cells in development: more than just noise. Development 136, 3853–3862 (2009)
Shalek, A. K. et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature 498, 236–240 (2013)
Shalek, A. K. et al. Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature 510, 363–369 (2014)
Ku, M. et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 4, e1000242 (2008)
Lee, T. I. et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125, 301–313 (2006)
Singer, Z. S. et al. Dynamic heterogeneity and DNA methylation in embryonic stem cells. Mol. Cell 55, 319–331 (2014)
Sigal, A. et al. Variability and memory of protein levels in human cells. Nature 444, 643–646 (2006)
Calabrese, J. M., Seila, A. C., Yeo, G. W. & Sharp, P. A. RNA sequence analysis defines Dicer’s role in mouse embryonic stem cells. Proc. Natl Acad. Sci. USA 104, 18097–18102 (2007)
Chamberlain, S. J., Yee, D. & Magnuson, T. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells 26, 1496–1505 (2008)
Kaji, K. et al. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nature Cell Biol. 8, 285–292 (2006)
Tsumura, A. et al. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells 11, 805–814 (2006)
Wang, Y., Medvid, R., Melton, C., Jaenisch, R. & Blelloch, R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nature Genet. 39, 380–385 (2007)
Tesar, P. J. et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448, 196–199 (2007)
Wang, Y. et al. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nature Genet. 40, 1478–1483 (2008)
Grün, D., Kester, L. & van Oudenaarden, A. Validation of noise models for single-cell transcriptomics. Nature Methods 11, 637–640 (2014)
Ebert, M. S. & Sharp, P. A. Roles for microRNAs in conferring robustness to biological processes. Cell 149, 515–524 (2012)
Leitch, H. G. et al. Naive pluripotency is associated with global DNA hypomethylation. Nature Struct. Mol. Biol. 20, 311–316 (2013)
Marks, H. et al. The transcriptional and epigenomic foundations of ground state pluripotency. Cell 149, 590–604 (2012)
Boyerinas, B., Park, S. M., Hau, A., Murmann, A. E. & Peter, M. E. The role of let-7 in cell differentiation and cancer. Endocr. Relat. Cancer 17, F19–F36 (2010)
Tsuruta, T. et al. miR-152 is a tumor suppressor microRNA that is silenced by DNA hypermethylation in endometrial cancer. Cancer Res. 71, 6450–6462 (2011)
Reinhart, B. J. et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403, 901–906 (2000)
Melton, C., Judson, R. L. & Blelloch, R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature 463, 621–626 (2010)
Zhu, H. et al. The Lin28/let-7 axis regulates glucose metabolism. Cell 147, 81–94 (2011)
Balázsi, G., van Oudenaarden, A. & Collins, J. J. Cellular decision making and biological noise: from microbes to mammals. Cell 144, 910–925 (2011)
Blake, W. J. et al. Phenotypic consequences of promoter-mediated transcriptional noise. Mol. Cell 24, 853–865 (2006)
Blake, W. J., Kaern, M., Cantor, C. R. & Collins, J. J. Noise in eukaryotic gene expression. Nature 422, 633–637 (2003)
Sommer, C. A. et al. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells 27, 543–549 (2009)
Park, I. H., Lerou, P. H., Zhao, R., Huo, H. & Daley, G. Q. Generation of human-induced pluripotent stem cells. Nature Protocols 3, 1180–1186 (2008)
Dow, L. E. et al. A pipeline for the generation of shRNA transgenic mice. Nature Protocols 7, 374–393 (2012)
Lee, S. H., Lumelsky, N., Studer, L., Auerbach, J. M. & McKay, R. D. Efficient generation of midbrain and hindbrain neurons from mouse embryonic stem cells. Nature Biotechnol. 18, 675–679 (2000)
Okabe, S., Forsberg-Nilsson, K., Spiro, A. C., Segal, M. & McKay, R. D. Development of neuronal precursor cells and functional postmitotic neurons from embryonic stem cells in vitro. Mech. Dev. 59, 89–102 (1996)
Ramsköld, D. et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nature Biotechnol. 30, 777–782 (2012)
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009)
Fujita, P. A. et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 39, D876–D882 (2010)
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011)
Patro, R., Mount, S. M. & Kingsford, C. Sailfish enables alignment-free isoform quantification from RNA-seq reads using lightweight algorithms. Nature Biotechnol. 32, 462–464 (2014)
Raj, A., van den Bogaard, P., Rifkin, S. A., van Oudenaarden, A. & Tyagi, S. Imaging individual mRNA molecules using multiple singly labeled probes. Nature Methods 5, 877–879 (2008)
Carpenter, A. E. et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7, R100 (2006)
Garber, M. et al. A high-throughput chromatin immunoprecipitation approach reveals principles of dynamic gene regulation in mammals. Mol. Cell 47, 810–822 (2012)
Spector, D. L. & Smith, H. C. Redistribution of U-snRNPs during mitosis. Exp. Cell Res. 163, 87–94 (1986)
Lewis, B. P., Burge, C. B. & Bartel, D. P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20 (2005)
Vejnar, C. E. & Zdobnov, E. M. MiRmap: comprehensive prediction of microRNA target repression strength. Nucleic Acids Res. 40, 11673–11683 (2012)
Holm, S. A simple sequentially rejective multiple test procedure. Scand. J. Stat. 6, 65–70 (1979)
Ashburner, M. et al. Gene ontology: tool for the unification of biology. Nature Genet. 25, 25–29 (2000)
Matthews, L. et al. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 37, 619–622 (2009)
Fraley, C. & Raftery, A. E. Model-based clustering, discriminant analysis, and density estimation. J. Am. Stat. Assoc. 458, 611–631 (2002)
Cahan, P. et al. CellNet: network biology applied to stem cell engineering. Cell 158, 903–915 (2014)
Acknowledgements
We thank members of the Collins and Daley laboratories for discussions. J.J.C. is supported by NIH grant R24DK092760 and the HHMI. G.Q.D. is supported by grants from the NIH (R01GM107536, R24DK092760, P50HG005550) and is an affiliate member of the Broad Institute and an investigator of the Manton Center for Orphan Disease Research and the Howard Hughes Medical Institute. A.R. is supported by the Broad Institute, the Klarman Cell Observatory at the Broad Institute, an NIH CEGS (1P50HG006193- 01), an NIH Pioneer Award (DP1OD003958-01) and the HHMI. R.M.K. is supported by the Wyss Institute. P.C. is supported by NIDDK (K01DK096013) and received support from NHLBI (T32HL066987 and T32HL007623) and the Manton Center for Orphan Disease Research. R.S. was supported by an NIH Postdoctoral Fellowship (1F32HD075541-01). H.L. is supported by the Mayo Clinic Center for Individualized Medicine. Sequencing was performed by the Broad Institute Genomics Platform. Flow cytometry was performed in the Hematologic Neoplasia Flow Cytometry Facility at the Dana-Farber Cancer Institute and the BCH IDDRC Stem Cell Core Facility at Boston Children’s Hospital supported by NIH-P30-HD18655. Single-cell qPCR experiments were performed at the BCH IDDRC Molecular Genetics Core Facility at Children’s Hospital Boston supported by NIH-P30-HD18655. Fludigm C1 experiments were performed at the Broad Institute and the Biopolymers Facility at Harvard Medical School.
Author information
Authors and Affiliations
Contributions
R.M.K. designed and performed the experiments, analysed the data and wrote the paper. P.C. analysed the data, developed analytical tools and wrote the paper. A.K.S., D.G. and J.J.T. performed ChIP-seq and single-cell RNA-seq experiments. R.S. helped to analyse the ChIP-seq and single-cell RNA-seq data. A.D. performed experiments and helped to analyse the data. H.L. helped to analyse the data. J.Z. generated the iShLin28 mESCs. K.P. generated the iPSCs. T.C.F. assisted with imaging and wrote image analysis algorithms. A.R. oversaw single-cell RNA-seq experiments and helped to write the paper. J.J.C. and G.Q.D. oversaw the project and helped to design the study and write the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Quality control of single-cell RNA-seq data.
a, A combination of read alignment rate (y axis) and number of genes detected (ln[TPM] >1) (x axis) was used to identify outlier cells (red circles) to remove from subsequent analysis, leaving 183 single mESCs cultured in serum+LIF, 94 mESCs cultured in 2i+LIF, and 84 Dgcr8−/− mESCs cultured in serum+LIF that were analysed by single-cell RNA-seq in this study (blue circles). b, Correlation in mean expression in detected cells (μ) between replicate serum+LIF plates across a range of α (fraction of cells a gene was detected in) thresholds. μ was calculated separately for each plate. Correlations in μ were calculated after limiting genes to those with α thresholds exceeding the specified threshold on the x axis. c, Maximum gene expression in replicate plates. Genes not reliably detected as defined in the text are coloured red. d, Comparison of α estimates in replicate plates, limited to reliably detected genes. Selected lineage regulators are denoted. e, Comparison of μ estimates in replicate plates, limited to reliably detected genes. Selected housekeeping, pluripotency and signalling genes are denoted. f, Top: relationship between estimates of α (x axis) and μ (y axis) of all genes based on both plates of mESC in FBS+LIF. Undetected genes are coloured yellow. Bottom: same as above except only showing reliably detected genes, and overlaid with density contour (red lines).
Extended Data Figure 2 Examination of mESCs cultured in serum+LIF for the presence of distinct subpopulations.
a, Hierarchical clustering dendrogram of single-cell RNA-seq data for 183 mESCs cultured in serum+LIF. b, Principal component analysis of the 183 mESCs cultured in serum+LIF. Points coloured blue are those with PCA1 values <−25, which are classified as outlier cells. c, Box plots of expression of selected pluripotency regulators and the lineage regulator Pax3 (top), and genes associated with EpiSCs (bottom). ‘Normal’ indicates the majority of cells coloured as grey dots in Extended Data Fig. 2b; ‘Outliers’ indicates the distinct set of 14 cells coloured blue in Extended Data Fig. 2b. P values for statistically significant differences are shown. d, Histograms showing the expression distributions of pluripotency regulators previously found to fluctuate within mESC populations. Cutoffs to divide expression into high and low states to test for enrichment within outlier cells are indicated by dashed lines. e, Average expression of Polycomb target genes within outlier cells (left), and the majority of the mESCs cultured in serum+LIF (right).
Extended Data Figure 3 Correlation of single-cell expression measurements between different technologies.
a, Correlation between transcripts per million (TPM) measured by single-cell RNA-seq and transcripts per cell measured by single-molecule FISH for 14 selected genes in mESCs cultured in serum+LIF. Error bars represent standard deviations of measurements. b, Correlation between coefficients of variation measured by single-cell RNA-seq and FISH for the 14 genes shown in panel a. c, Correlation coefficients for α (fraction detected, left) or μ (mean expression in detected cells, right) between single-molecule FISH and single-cell RNA-seq are plotted as a function of varying the threshold level for detection by RNA FISH (x axis). An RNA FISH detection threshold of 10 indicates that genes expressed at <10 copies per cell would not be detected by RNA FISH. Correlation for α between RNA-seq and RNA FISH peaked at an RNA FISH detection threshold of >5 transcripts per cell, giving an estimated single-cell RNA-seq detection efficiency of ∼20% (1 out of 5 transcripts detected, assuming single-molecule sensitivity for the RNA FISH method). d, Correlation in α between single-cell RNA-seq and single-molecule FISH for 14 genes measured by both methods, assuming a single-molecule FISH detection threshold of >5 transcripts per cell. Dashed line shows linear fit to the data. The fraction of cells a gene is detected in shows good agreement between the two methods when taking the sensitivity of the RNA-seq into account. e, Comparison of the fraction of mESCs cultured in serum+LIF a gene was detected in by single-cell qPCR (x axis) or single-cell RNA-seq (y axis). Single-cell RNA-seq showed greater sensitivity overall as compared to single-cell qPCR, but a set of genes was sporadically expressed as measured by both methods. Trend line indicates linear fit to the data. f, Correlations of fraction detected between independent biological replicates for 96 genes profiled by single-cell qPCR. Trend line shows linear fit to the data, and indicates that the fraction of cells a gene is detected in remains consistent across independent biological replicates. g, Comparison of expression distributions measured by single-cell RNA-seq (light grey) and single-molecule FISH (darker grey) for pluripotency regulators (top) and Polycomb target genes (bottom).
Extended Data Figure 4 Expression of Polycomb target genes in ESCs and neural precursor cells (NPCs).
a, Polycomb target genes show highly variable expression in mESCs. Relationship between μ (mean expression in detected cells, x axis) and coefficient of variation (standard deviation normalized by mean expression in all cells, y axis) is shown for Polycomb target genes (purple) and non-Polycomb target genes (grey). Distributions for μ and coefficients of variation for the two gene sets are shown above and to the left of the graph, respectively. Polycomb target genes show pronounced variability in expression, even when controlling for expression level. b, Expression of the neural regulators and Polycomb target genes Otx2 (top) and Neurod1 (bottom) measured by RNA FISH in mESCs cultured in serum+LIF. Overall distributions within the population (left) and representative colonies (right) are shown, along with gene tracks from the Broad Institute Integrated Genomics Viewer (IGV) showing ChIP-seq reads for H3K27me3 at the Otx2 and Neurod1 genes. c, Enriched gene ontology categories among genes significantly upregulated (red) or downregulated (green) in neural precursor cells as compared to embryonic stem cells. d, Expression changes of selected genes in neural precursor cells as compared to the embryonic stem cells they were derived from. As expected, neural regulators and ES Polycomb target genes Pax6, Prrx1, Hes1, Msx1, Pbx3, Nes and Runx1 were upregulated in NPCs, while the pluripotency regulators Esrrb, Nr0b1, Pou5f1 (Oct4), Zfp42 (Rex1) and Nanog were downregulated. Expression of the housekeeping genes Gapdh and Actb, and the pluripotency and neural regulator Sox2, were relatively unchanged between the two cell types. e, Expression comparison of ES Polycomb target genes that are detected in either mESCs or NPCs. Many Polycomb target genes that are neuronal regulators are detected in a higher fraction of NPCs than ESCs, while certain Polycomb targets such as Pax3 (a regulator of musculoskeletal development) are detected in a smaller fraction of cells. Genes are ordered in ascending order of ESC to NPC average expression change. f, Histograms showing distributions of expression levels for selected housekeeping genes (grey) and neuronal regulators (green and red) in NPCs. The neuronal regulators Msx1 and Runx1 (red) show bimodal expression in NPCs. g, Distance distributions within ESC (red) and NPC (blue) populations. NPCs show more population heterogeneity than ESCs. h, State classification based on principal component analysis of single-cell RNA-seq of NPCs. Four distinct states are identified.
Extended Data Figure 5 Fluctuations in pluripotency and lineage regulator expression.
a, RNA FISH images showing expression of Nanog (top), Nr0b1 (middle) and Esrrb (bottom) in individual colonies or regions of cells. b, c, Histograms show distributions of fluorescence intensities within individual cells from quantitative immunofluorescence of OCT4 and (b) NANOG, or (c) NR0B1, along with NANOG/OCT4 or NR0B1/OCT4 ratios as indicated. For NANOG/OCT4 images (b), NANOG staining is coloured green while OCT4 staining is coloured red. In the panel on the left a cluster of low NANOG cells is indicated by a blue arrow, while a cluster of high NANOG cells is indicated by a white arrow. In the panel on the right, the same image is shown with DAPI staining coloured blue, and groups of OCT4 negative/NANOG negative differentiated cells that may have arisen from the low NANOG cells are indicated with grey arrows. For NR0B1/OCT4 images (c), NR0B1 is coloured green while OCT4 staining is coloured red. A relatively high NR0B1 colony is shown in the panel on the left, while a region of low NR0B1 cells is displayed in the panel on the right. d, Quantitative immunofluorescence images showing expression of OCT4 (red) and NR0B1 (green, top row), NANOG (green, middle row), or ESRRB (green, bottom row) within individual colonies of mESCs grown in serum+LIF. OCT4 is used as an internal reference as it shows relatively invariant expression within mESCs. e, Single-cell RNA-seq relationships for gene pairs shown in Fig. 2g. Distributions of gene expression from RNA-seq and RNA FISH experiments are shown. Dashed lines indicate divisions between high and low states for box plot shown in Fig. 2g. Single-cell RNA-seq data show that the subset of cells in both a high Sox2 and low Nr0b1 state show an increased probability of expressing Neurod1 (ln[TPM] >1) as compared to all cells (bottom). f, RNA-seq (left) and RNA FISH (right) correlations between the pluripotency regulator Esrrb and the signalling factor Bmp4 (top), and the pluripotency regulator Nanog and the neural regulator Otx2 (bottom). Dashed lines indicate divisions between high and low states for the box plots shown on the right. g, Correlations from single-cell RNA-seq data between average Polycomb target gene expression and Zfp42 (Rex1), Nanog and Nr0b1. Dashed lines indicate divisions between high and low states for the box plot comparing Polycomb target expression with expression of the three regulators, which are all negatively associated with expression of Polycomb targets. The Venn diagram shows coupling between high and low states of the three regulators. Low Nr0b1 cells are more likely to be in a low Zfp42 and/or low Nanog state as compared to high Nr0b1 cells, suggesting that Nr0b1 functions to maintain Zfp42 and Nanog expression and repress Polycomb target genes. h, RNA FISH images of an ESC colony hybridized with probes against Nanog (yellow) and Otx2 (magenta), showing inverse relationship between Nanog and Otx2 expression.
Extended Data Figure 6 Single-cell qPCR of mESCs exposed to chemical and genetic perturbations.
a, Shown are the average Ct values and standard deviations for technical triplicates (error bars on x axis) or biological triplicates (error bars on y axis) across 96 genes for pools of 100 or 10 cells or single sorted cells. Cells were sorted into PCR strips containing RT–PCR reagents and primer pools, reverse transcription and pre-amplification was performed, and cDNA was quantified on a Fluidigm BioMark PCR system. b, Heat map of single-cell qPCR data for 84 genes examined across 19 different PSC perturbations and in MEFs (n = 1,144 single cells). Unsupervised hierarchical clustering grouped genes into three clusters: bimodally expressed genes (right group), ubiquitously expressed genes (left group), and sporadically expressed genes (middle group). c, Numbers of genes showing significant changes in expression distributions as compared to the reference conditions of v6.5 mESCs cultured in serum+LIF on MEFs. Significance of changes was determined by the two-sample Kolmogorov–Smirnov test, correcting P values for multiple tests using Holm’s method. d, Selection of the state classification model that maximizes the Bayesian Information Criteria (BIC, y axis). Mclust was used to generate multivariate Gaussian mixture models of the first three principal components of the Fluidigm qPCR-based expression values of individual mESCs. The models vary in the number of components (one to nine) and the following geometric characteristics: volume, shape and orientation as described68. The best model was used to classify cells into states.
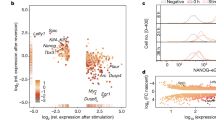
Extended Data Figure 7 Gene expression changes in mESCs upon culture in 2i or removal of mature miRNAs.
a, Changes in expression of the pluripotency regulators shown in Fig. 4c going from serum+LIF to 2i+LIF culture (x axis), or between wild-type and Dgcr8−/− cells cultured in serum+LIF (y axis), as measured by single-cell RNA-seq. Selected genes are highlighted. b, Changes in expression of 18 commonly used housekeeping genes (ActB, Aip, Cxxc1, Gapdh, Gusb, Hmbs, Hprt, Ipo8, Mrpl48, Mtcp1, Pgk1, Ppia, Rpl13a, Rplp2, Rps6, Tbp, Ubc and Ywhaz) between the same conditions as in panel a. c, Intra-condition (left) and inter-condition (right) distances between individual cells based on single-cell RNA-seq data for all genes (top), 219 transcription factors that regulate pluripotent cells as determined by CellNet69 (middle), or lineage regulators, defined as the 256 previously determined Polycomb target genes in mESCs that are transcription factors (bottom). d, Comparison of single-cell average expression changes going from serum+LIF to 2i+LIF culture in the present study (x axis) against population-level expression changes between mESCs cultured in serum+LIF versus 2i+LIF measured by ref. 40 (y axis). Trend line from linear fit to the data are shown, and selected genes that show lower expression in 2i+LIF culture in both studies are highlighted. e, Single-molecule FISH showing shifts in expression of Oct4, Nanog, Nr0b1 and Otx2 at the RNA level between wild-type mESCs in serum+LIF and 2i+LIF culture conditions and Dgcr8−/− mESCs cultured in serum+LIF. f, Representative RNA FISH images showing expression of the Polycomb target gene and neural regulator Otx2 in individual mESC colonies under the three conditions examined. g, Correlation between expression shifts between serum+LIF and 2i+LIF culture observed by single-cell RNA-seq (x axis) and RNA FISH (y axis) for the 14 genes shown in Extended Data Fig. 3a. Trendline indicates linear fit to the data. h, Quantitative immunofluorescence showing changes in NANOG/OCT4 (left) and NR0B1/OCT4 (right) ratios between serum+LIF and 2i+LIF culture. Serum+LIF data are the same shown in Extended Data Fig. 5. i, RNA FISH images of E14 mESC colonies cultured in serum+LIF (left) or 2i+LIF (right) media and probed for Nanog (top) or Esrrb (bottom) expression. Both Nanog and Esrrb show bimodal expression patterns in E14 mESCs grown in serum+LIF, and shift towards the high expression state in 2i+LIF culture. White arrows indicate regions of low Nanog or Esrrb expression in mESCs grown in serum+LIF.
Extended Data Figure 8 Dependence of Polycomb target gene expression on culture conditions and miRNAs.
a, Fraction of Polycomb target genes detected in wild-type mESCs cultured in serum+LIF (red) or 2i+LIF (green), or Dgcr8−/− mESCs cultured in serum+LIF (blue). b, Correlation between Polycomb target gene expression and pluripotency regulator expression in different conditions (serum+LIF in blue, Dgcr8−/− in orange, and 2i+LIF in red). Displayed are the Pearson correlation coefficients (PCC) between pluripotency-related regulator z-score and proportion of Polycomb targets detected, computed across all single cells. The z-score is defined as the number of standard deviations that a sample exceeds (z-score >0) or is less than (<0) the mean value. z-scores for pluripotency regulators were computed for each condition separately. A low PCC indicates that a lower factor expression (relative to its mean in the condition) increases the likelihood that Polycomb targets will be detected as expressed (for example, Zfp42 in FBS+Lif). c, Scatter plots comparing amount of H3K9me3 (top), H3K27ac (middle) and RNA polymerase II (bottom) at promoter regions in wild-type mESCs cultured in serum+LIF versus 2i+LIF conditions, wild-type mESCs versus Dgcr8−/− mESCs cultured in serum+LIF, or Dgcr8−/− mESCs cultured in serum+LIF versus wild-type mESCs cultured in 2i+LIF as indicated. ChIP-seq reads at gene promoters were median normalized for comparison, and Polycomb target genes are indicated in green. Unlike H3K27me3, levels of these three factors do not show a strong decrease at Polycomb target genes under 2i+LIF conditions and in Dgcr8−/− mESCs (compare to Fig. 4f).
Extended Data Figure 9 Perturbing miRNA balance and the Myc/Lin28/let-7 axis.
a, Purified RNA from v6.5 mESCs and E14 mESCs cultured in either serum+LIF or 2i+LIF conditions was extracted and reverse-transcribed with TaqMan primers specific to the indicated miRNAs, and then expression was profiled by qPCR. Error bars represent standard deviation from technical triplicate PCR reactions, and samples are normalized to a basket of reference small noncoding RNAs and the decrease in Ct values compared to v6.5 mESCs grown in serum+LIF is shown. See Methods for full details. b, Fold-change in expression of the indicated miRNAs in wild-type mESCs cultured in serum+LIF or 2i+LIF, induced and uninduced ilet-7 mESCs cultured in serum+LIF, and MEFs grown under standard conditions. Changes are shown relative to v6.5 mESCs grown in serum+LIF. c, Comparison of genes that change in expression upon introduction of the ESCC miRNA miR-294 to Dgcr8−/− mESCs in ref. 44 (y axis) to genes that change in expression between serum+LIF and 2i+LIF culture in single-cell RNA-seq data (x axis). Genes that are significantly upregulated in Dgcr8−/− mESCs upon miR-294 introduction are indicated in blue, and those that are significantly downregulated are indicated in red. Selected genes upregulated by miR-294 and downregulated in 2i+LIF culture as compared to serum+LIF are highlighted. As a group, genes downregulated by miR-294 show higher expression in 2i+LIF than in serum+LIF. d, mESC colonies staining uniformly positive for alkaline phosphatase show reduced levels of Myc. Overall and colony-specific Myc distribution in serum+LIF culture as measured by RNA FISH, showing uniformly positive (top), mixed (middle), or negative (bottom) alkaline phosphatase staining. e, Western blot showing reduction of Lin28a protein levels in iShLin28a cells upon addition of doxycycline. f, let-7 expression changes in doxycycline-inducible ilet-7 mESCs grown in serum+LIF upon induction. RT–qPCR was performed as in panel a, and Ct changes are shown relative to v6.5 mESCs in serum+LIF in a separate experiment from panel a. The inducible let-7 construct is detected by the let-7g probe. g, Effect of miRNA transfection on self-renewal efficiency of Dgcr8−/− mESCs. miRNA mimics were transfected into Dgcr8−/− mESCs, and self-renewal efficiency was measured. Error bars indicate standard deviations between triplicate transfection experiments. Co-transfection of the ESCC miRNA miR-294 with a let-7 miRNA results in enhanced self-renewal efficiency as compared to miR-294 alone. h, Expression changes of selected genes measured by qPCR upon culture of v6.5 mESCs in serum+LIF, 2i+LIF, or treatment with only Erk or GSK3β inhibitors.
Supplementary information
Supplementary Information
This file contains the Supplementary Discussion and Supplementary References. (PDF 491 kb)
Supplementary Table 1
Single-cell and population RNA-Seq expression values of wild type (v6.5) mouse ESCs cultured in FBS and LIF. This file contains single-cell RNA-Seq expression values, outlier status from Extended Data Figure 2, and sequencing metrics for mESCs cultured in serum+LIF. (XLSX 27967 kb)
Supplementary Tables 2-4
This zipped file contains Supplementary Tables 2-4. Supplementary Table 2 contains single-cell and population RNA-Seq expression value statistics of wild type (v6.5) mouse ESCs cultured in either FBS and LIF, or 2i and LIF, or of DGCR8-/- mouse ESCs cultured in FBS and LIF. Supplementary Table 3 contains expression values and statistics for non-coding RNAs in single-cell RNA-Seq data from mESCs are listed. Supplementary Table 4 contains expression value statistics from single-cell RNA-Seq profiling of ESC-derived neural precursor cells. (ZIP 26403 kb)
Supplementary Tables 5-7
This zipped file contains Supplementary Tables 5-7. Supplementary Table 5 contains ChIP-Seq reads from H3K4me3, RNAP2, H3K9me3, H3K27me3, H3K36me3, and H3K27ac immunoprecipitations in wild type (v6.5) mESC cultured in FBS and LIF, or in 2i and LIF, or DGCR8-/- mESCs cultured in FBS and LIF. The Number of reads overlapping with the promoter of each gene (from -1.5kb upstream to 0.5kb downstream of the transcription start site) are listed. Supplementary Table 6 contains single-cell qPCR expression data. Ct values for the single-cell qPCR data shown in Figure 3 are listed, along with state classifications for individual cells, PCA loadings for the each gene, and TaqMan probes used in the study. Supplementary Table 7 contains single-cell and population RNA-Seq expression values of wild type (v6.5) mouse ESCs cultured in 2i and LIF. (ZIP 21142 kb)
Supplementary Tables 8-10
This zipped file contains Supplementary Tables 8-10. Supplementary Table 8 contains single cell and population RNA-Seq expression values of DGCR8-/- mouse ESCs cultured in FBS and LIF. Supplementary Table 9 contains RNA-seq single cell based differential gene expression between mESC cultured in FBS and LIF, in 2i and LIF, or DGCR8-/- mESC cultured in FBS and LIF. Nominal p-values, Students t-test statistic, mean expression values, Holms corrections, and false discovery rates are given for differential gene expression comparisons between the three mESC populations profiled by single-cell RNA-Seq. Supplementary Table 10 contains NanoString miR expression of mouse embryonic fibroblasts (MEFs), v6.5 and E14 mESCs cultured in FBS and LIF, or in 2i and LIF, and DGCR8-/- cultured in FBS and LIF. Expression values from NanoString miRNA profiling of the indicated populations are listed. (ZIP 19168 kb)
Rights and permissions
About this article
Cite this article
Kumar, R., Cahan, P., Shalek, A. et al. Deconstructing transcriptional heterogeneity in pluripotent stem cells. Nature 516, 56–61 (2014). https://doi.org/10.1038/nature13920
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13920
This article is cited by
-
Gene expression model inference from snapshot RNA data using Bayesian non-parametrics
Nature Computational Science (2023)
-
Diagnostic classification of childhood cancer using multiscale transcriptomics
Nature Medicine (2023)
-
From nasal to basal: single-cell sequencing of the bursa of Fabricius highlights the IBDV infection mechanism in chickens
Cell & Bioscience (2021)
-
Pluripotent stem cell-derived models of neurological diseases reveal early transcriptional heterogeneity
Genome Biology (2021)
-
Repression of germline genes by PRC1.6 and SETDB1 in the early embryo precedes DNA methylation-mediated silencing
Nature Communications (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.