Abstract
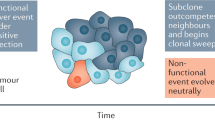
Cancers arise through a process of somatic evolution that can result in substantial sub-clonal heterogeneity within tumours. The mechanisms responsible for the coexistence of distinct sub-clones and the biological consequences of this coexistence remain poorly understood. Here we used a mouse xenograft model to investigate the impact of sub-clonal heterogeneity on tumour phenotypes and the competitive expansion of individual clones. We found that tumour growth can be driven by a minor cell subpopulation, which enhances the proliferation of all cells within a tumour by overcoming environmental constraints and yet can be outcompeted by faster proliferating competitors, resulting in tumour collapse. We developed a mathematical modelling framework to identify the rules underlying the generation of intra-tumour clonal heterogeneity. We found that non-cell-autonomous driving of tumour growth, together with clonal interference, stabilizes sub-clonal heterogeneity, thereby enabling inter-clonal interactions that can lead to new phenotypic traits.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Greaves, M. & Maley, C. C. Clonal evolution in cancer. Nature 481, 306–313 (2012)
Nowell, P. C. The clonal evolution of tumor cell populations. Science 194, 23–28 (1976)
Fearon, E. R. & Vogelstein, B. A genetic model for colorectal tumorigenesis. Cell 61, 759–767 (1990)
Gerlinger, M. et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 366, 883–892 (2012)
Landau, D. A. et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell 152, 714–726 (2013)
Ding, L. et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 481, 506–510 (2012)
Burrell, R. A., McGranahan, N., Bartek, J. & Swanton, C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature 501, 338–345 (2013)
Anderson, K. et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature 469, 356–361 (2011)
Marusyk, A. & Polyak, K. Tumor heterogeneity: causes and consequences. Biochim. Biophys. Acta 1805, 105–117 (2010)
Merlo, L. M., Pepper, J. W., Reid, B. J. & Maley, C. C. Cancer as an evolutionary and ecological process. Nature Rev. Cancer 6, 924–935 (2006)
Weinberg, R. A. The Biology of Cancer (Garland Science, 2007)
Bissell, M. J. & Hines, W. C. Why don't we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nature Med. 17, 320–329 (2011)
Gatenby, R. A. & Gillies, R. J. A microenvironmental model of carcinogenesis. Nature Rev. Cancer 8, 56–61 (2008)
DeGregori, J. Challenging the axiom: does the occurrence of oncogenic mutations truly limit cancer development with age? Oncogene 32, 1869–1875 (2013)
Kreso, A. et al. Variable clonal repopulation dynamics influence chemotherapy response in colorectal cancer. Science 339, 543–548 (2013)
Gerrish, P. J. & Lenski, R. E. The fate of competing beneficial mutations in an asexual population. Genetica 102–103, 127–144 (1998)
Putoczki, T. & Ernst, M. More than a sidekick: the IL-6 family cytokine IL-11 links inflammation to cancer. J. Leukoc. Biol. 88, 1109–1117 (2010)
Ernst, M. et al. STAT3 and STAT1 mediate IL-11-dependent and inflammation-associated gastric tumorigenesis in gp130 receptor mutant mice. J. Clin. Invest. 118, 1727–1738 (2008)
Putoczki, T. L. et al. Interleukin-11 is the dominant IL-6 family cytokine during gastrointestinal tumorigenesis and can be targeted therapeutically. Cancer Cell 24, 257–271 (2013)
Inda, M. M. et al. Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes Dev. 24, 1731–1745 (2010)
Bissell, M. J. & Radisky, D. Putting tumours in context. Nature Rev. Cancer 1, 46–54 (2001)
Folkman, J. Angiogenesis in cancer, vascular, rheumatoid and other disease. Nature Med. 1, 27–31 (1995)
Levayer, R. & Moreno, E. Mechanisms of cell competition: themes and variations. J. Cell Biol. 200, 689–698 (2013)
Maley, C. C., Reid, B. J. & Forrest, S. Cancer prevention strategies that address the evolutionary dynamics of neoplastic cells: simulating benign cell boosters and selection for chemosensitivity. Cancer Epidemiol. Biomarkers Prev. 13, 1375–1384 (2004)
Nagy, J. D. Competition and natural selection in a mathematical model of cancer. Bull. Math. Biol. 66, 663–687 (2004)
Wu, M., Pastor-Pareja, J. C. & Xu, T. Interaction between RasV12and scribbled clones induces tumour growth and invasion. Nature 463, 545–548 (2010)
Cleary, A. S., Leonard, T. L., Gestl, S. A. & Gunther, E. J. Tumour cell heterogeneity maintained by cooperating subclones in Wnt-driven mammary cancers. Nature 508, 113–117 (2014)
Calbo, J. et al. A functional role for tumor cell heterogeneity in a mouse model of small cell lung cancer. Cancer Cell 19, 244–256 (2011)
Navin, N. E. & Hicks, J. Tracing the tumor lineage. Mol. Oncol. 4, 267–283 (2010)
Preibisch, S., Saalfeld, S. & Tomancak, P. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics 25, 1463–1465 (2009)
Acknowledgements
We thank J. DeGregori, A. Goldman, A. Rozhok, M. Gonen and members of the Polyak and Michor laboratories for their critical reading of this manuscript and discussions. We thank L. Cameron in the DFCI Confocal Microscopy for her technical support. This work was supported by the Dana-Farber Cancer Institute Physical Sciences-Oncology Center (U54CA143798 to F.M.), CDRMP Breast Cancer Research Program W81XWH-09-1-0561 (A.M.), Cellex Foundation (V.A.), Deutsche Akademie der Naturforscher Leopoldina LPDS 2012-12 (P.M.A.) and the Breast Cancer Research Foundation (K.P.).
Author information
Authors and Affiliations
Contributions
A.M. developed the experimental model, performed xenograft experiments and data analyses. D.P.T. performed immunohistochemical analyses and quantifications, and assisted with animal experiments. P.M.A. performed mathematical modelling and data analyses. V.A. assisted with image acquisition and analyses. K.P. supervised with help from F.M. All authors helped to design the study and write the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Proliferation, apoptosis and vascularization in selected groups.
a–c, Quantification and representative pictures of immunohistochemical analysis for markers of proliferation (a), apoptosis (b), and vascularization (c). Each dot represents an individual tumour, error bars indicate s.d.
Extended Data Figure 2 Estimations of clonal frequencies.
a, Schematic outline of the quantification of clonal composition based on qPCR. Changes in clonal frequencies are determined based on changes in the ratios of clone-specific and a human-specific reference amplicon between initial mixtures and the resulting tumours. b, Reproducibility of clonality analysis between two different DNA preparations/qPCR from same tumour. c, Correlation between the results obtained using fluorescent-activated cell sorting (FACS) and qPCR based determination of clonal frequency after 6 weeks in vitro culture. Green fluorescent protein (GFP) labelled parental cells were mixed with individual sub-clones at initial ratios of 20:1. R2 indicates goodness of fit of linear regression.
Extended Data Figure 3 Mathematical model.
a, Upper panel: estimation of tumour volume–density relation. The dashed line represents a linear regression with slope 0.33 (P < 0.01). Red dots are predictions for which one value of the pair was missing. Inset, tumour density over time from clone-vs-parental competition experiments (dots). Tumour density did not correlate with the time of sample collection (line, linear regression with slope 0.012, P = 0.68). Lower panel, schematic of estimation of cell numbers in tumour samples from two dimensional slices. b, Tumour volume over time from experiments (empty circles) and linear regression (exponential tumour growth law, black lines), with 0.95 confidence intervals (grey areas). Inset: comparison of P values using different growth laws. c, Flow chart of mathematical modelling approach. d, Upper panel, growth dynamics under non-cell-autonomous driving, according to mathematical model (model B, see Supplementary Information), driver effect of IL11 was set to a typical value of 0.012/day. Example of four individual sub-clones (for example, IL11, LOXL3, slow-growing CCL5, LacZ), total tumour size indicated by dashed line; lower panel, frequency dynamics for the same set.
Extended Data Figure 4 Reproducibility and frequency-independence of tumour-growth promoting effects of IL11.
a, Relation between tumour weight and fraction of IL11 sub-clone cells upon tumour sample collection. b, Final weights of tumours initiated from the indicated mixtures of IL11 expressing and parental cells using pLenti6.3 backbone; n = 21 for the 5.6% IL11, n = 10 for the other groups. c, d, Secreted (pg per 106 cells per hour) (c) and intracellular (pg per 106 cells) (d) levels of IL11 protein determined by ELISA in parental cells and in the IL11-expressing clones derived using the indicated lentiviral constructs. e, Growth kinetics of tumours initiated by transplantation of mixtures containing IL11-expressing cells from the indicated backbones competing with the parental cells.
Extended Data Figure 5 IL11 in clonal cooperation.
a, Expansion (fold-change over initial number of cells) of indicated sub-clones in the polyclonal tumours initiated with/without IL11 sub-clone, n = 10 per group. b, Growth curves of the tumours initiated by transplantation of the indicated groups, IL11 + FIGF indicates tumours initiated by 1:1 mixtures of IL11 and FIGF sub-clones.
Extended Data Figure 6 The effects of doxorubicin on tumour growth and clonal composition.
a–c, Tumour growth (a), assessment of cell proliferation by BrdU staining (b) and clonal composition (c) of tumours initiated by polyclonal mixtures followed by treatment of the animals bearing established tumours with vehicle control or doxorubicin. Arrows mark intraperitoneal injections of doxorubicin (5 mg per kg) or vehicle. The inset in c quantifies changes in frequency of clones expanding and shrinking compared to the initial frequencies. Interaction factor for two-way ANOVA between control and doxorubicin groups is statistically significant (P = 0.0059). d, Shannon index for clonal diversity of vehicle and doxorubicin treated tumours, *P < 0.05 in two-sample Kolmogorov–Smirnov test.
Extended Data Figure 7 Validation of IL11Rα shRNA.
As the commercially available IL11Rα antibodies are not sufficiently sensitive to detect endogenous IL11Rα protein in the MDA-MB-468 cells, we tested the ability of shRNA to downregulate the expression of exogenously expressed IL11Rα. Cells overexpressing IL11Rα were stably transduced with IL11Rα-targeting shRNAs and the expression of IL11Rα and β-actin (loading control) were analysed by immunoblotting.
Extended Data Figure 8 The effects of IL11 on the tumour microenvironment.
a, Collagen organization in parental and IL11 expressing tumours. Representative images of collagen structure (blue) in the indicated tumours as determined by tri-chrome staining. b, Smooth muscle actin positive (SMA) stromal cells in control and IL11 expressing tumours. Representative images of immunohistochemical staining for SMA.
Extended Data Figure 9 IL11 cells are not specifically eliminated in IL11/LOXL3 tumours.
a, Immunofluorescence analysis of apoptosis in 1:1 IL11/LOXL3 tumours. Apoptotic marker cleaved caspase 3 (yellow) indicates lack of increase in apoptosis in IL11 (red, V5+) cells bordering LOXL3 (V5−, as LOXL3 cDNA has a stop codon before the tag). Grey dashed line demarcates the border of the necrotic area, where most of cell death occurs. b, Occasional IL11+ cells (indicated by arrows) could still be detected in the remnants of 1:18 IL11/LOXL3 tumours.
Supplementary information
Supplementary Information
This file contains a Mathematical Supplement, Supplementary References and Supplementary Tables 1-6. (PDF 382 kb)
Rights and permissions
About this article
Cite this article
Marusyk, A., Tabassum, D., Altrock, P. et al. Non-cell-autonomous driving of tumour growth supports sub-clonal heterogeneity. Nature 514, 54–58 (2014). https://doi.org/10.1038/nature13556
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13556
This article is cited by
-
The relationship between high ratios of CD4/FOXP3 and CD8/CD163 and the improved survivability of metastatic triple-negative breast cancer patients: a multicenter cohort study
BMC Research Notes (2024)
-
Extensive intratumor regional epigenetic heterogeneity in clear cell renal cell carcinoma targets kidney enhancers and is associated with poor outcome
Clinical Epigenetics (2023)
-
Intratumour heterogeneity, from hypothesis to the clinic
British Journal of Cancer (2023)
-
State-dependent evolutionary models reveal modes of solid tumour growth
Nature Ecology & Evolution (2023)
-
Identifying the effectiveness of 3D culture systems to recapitulate breast tumor tissue in situ
Cellular Oncology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.