Abstract
We conducted a genome-wide association study for nonsyndromic cleft lip with or without cleft palate (NSCL/P) in 401 affected individuals and 1,323 controls, with replication in an independent sample of 793 NSCL/P triads. We report two new loci associated with NSCL/P at 17q22 (rs227731, combined P = 1.07 × 10−8, relative risk in homozygotes = 1.84, 95% CI 1.34–2.53) and 10q25.3 (rs7078160, combined P = 1.92 × 10−8, relative risk in homozygotes = 2.17, 95% CI 1.32–3.56).
Similar content being viewed by others
Main
NSCL/P is one of the most common human birth defects. In European populations, NSCL/P has a prevalence ranging from 1 in 700 to 1 in 1,000. We recently reported a susceptibility locus for NSCL/P at chromosome 8q24.21 from a genome-wide association study in 224 individuals with NSCL/P (cases) and 383 population-based controls1. This locus is the second susceptibility locus to have been unequivocally identified for NSCL/P to date, the first being the IRF6 locus2.
To identify additional cleft susceptibility loci, we enlarged our sample by genotyping an additional set of 177 NSCL/P cases and adding the genotypes of 940 population-based controls of central European origin. Genotyping was performed using Illumina BeadChips (Human610-Quad and HumanHap 550k).
Following quality control (Supplementary Methods and Supplementary Fig. 1), association analysis of 521,288 SNPs having a minor allele frequency (MAF) of ≥1% in controls was performed in 399 cases and 1,318 controls.
After excluding markers from the previously described 8q24.21 locus, 20 SNPs with P < 10−5 remained. Five chromosomal loci (8q12.3, 10q25.3, 13q31.1, 15q13.3 and 17q22) were located within these 20 top SNPs, and the associations at these loci were further supported by at least three more SNPs with P < 10−4 (Supplementary Fig. 2 and Supplementary Table 1). Two additional regions were considered to be promising NSCL/P susceptibility loci (6p22.1, 11q14.2), as they contained at least four markers with P < 10−4.
To replicate the genome-wide association study (GWAS) findings, we selected the 20 top SNPs (P < 10−5) as well as additional backup markers for each of the seven previously mentioned loci, resulting in two replication assays. We included additional SNPs with P < 10−4 in the two replication assays, giving highest priority to SNPs with the lowest P values. Thus, a total of 56 markers were genotyped in a replication sample of 793 NSCL/P triads of European origin. Genotyping using matrix assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (Sequenom Inc.) was successful for 45 markers (representing 32 different loci), which were then analyzed by the transmission-disequilibrium test in 665 triads (128 triads were excluded after quality control, Supplementary Methods).
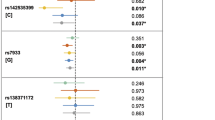
Of the 45 SNPs successfully genotyped, 11 (representing six different loci) showed P < 0.05 in the replication sample (Supplementary Table 2). Two of these SNPs remained significant after correction for multiple testing by a conservative Bonferroni procedure (17q22: rs227731, Pcorr = 0.01039 and 10q25.3: rs7078160, Pcorr = 0.04999). The probability that 6 or more out of 32 loci would generate P values <0.05 by chance alone is 0.0046. It is therefore likely that true association was detected. After combining the GWAS and replication samples, genome-wide significant evidence of association was found using the combined haplotype relative risk method3 for three SNPs at two loci (17q22: rs227731, Pcomb = 1.07 × 10−8; and 10q25.3: rs7078160, Pcomb = 1.92 × 10−8 and rs4752028, Pcomb = 2.48 × 10−8) (Table 1, Supplementary Table 3 and Supplementary Methods). Two further loci (13q31.1, 15q13.3) were replicated, although they fell short of achieving genome-wide significance (13q31.1: rs9574565, Pcomb = 3.44 × 10−7 and 15q13.3: rs1258763, Pcomb = 1.14 × 10−6). Similarly, the single marker rs7590268 on 2p21 failed to reach genome-wide significance in the combined sample (Pcomb = 8.62 × 10−8). For the two significant markers on 6q27 (rs2197100 and rs7740603, Supplementary Table 2), the putative risk alleles in the replication study differed from those in the GWAS, and these markers were therefore not considered to have been replicated. There was no evidence for imprinting or a maternal genotype effect for any of the 45 SNPs included in the family-based replication step (Supplementary Methods).
The relative risk (RR) in the replication sample for rs227731 (17q22) was 1.26 (95% CI 0.99–1.6) for the heterozygous genotype and 1.84 (95% CI 1.34–2.53) for the homozygous genotype. The RR in the replication sample for rs7078160 (10q25.3) was 1.32 (95% CI 1.05–1.65) for the heterozygous genotype and 2.17 (95% CI 1.32–3.56) for the homozygous genotype (Table 1).
The population attributable risk (PAR) estimated from the combined sample was 23.9% for rs227731 (17q22) and 12.3% for rs7078160 (10q25.3). The joint PAR for these two new loci, the key susceptibility locus at 8q24.21 and the IRF6 locus estimated from the combined sample was 54.6% (Supplementary Methods). Although there may be biases in estimating the PAR from these discovery cohort samples, these results suggest that together these four loci may explain a substantial proportion of the risk for NSCL/P. However, because the summary PAR is usually less than the sum of the individual PARs, it is also likely that additional genetic variants contribute to NSCL/P risk and remain to be identified.
The chromosome 17q22 region contains the gene encoding the noggin protein, NOG, which is located 100 kb centromeric of rs227731 (Fig. 1a). NOG is an antagonist of members of the transforming growth factor beta (TGF-β) superfamily, which includes proteins such as the bone morphogenetic protein 4 (BMP4). BMP4 has been shown to regulate mammalian palatogenesis4 and has been reported to be associated with clefting in humans5.
(a,b) Each panel shows single-marker association statistics (as −log10 P; left y axis) from the GWAS (squares) and from the combined analysis (diamonds). Linkage disequilibrium (r2) to the most significantly associated regional SNP (blue diamond), as estimated from the GWAS control genotypes, is color-coded (red: r2 > 0.8; orange: r2 = 0.5–0.8; yellow: r2 = 0.2–0.5; white: r2 < 0.2). Recombination rates across each region in HapMap CEU are shown in light blue (right y axis). The chromosomal locations and relative positions of genes according to hg18 are shown (x axis).
The four markers at 10q25.3 with a PGWAS < 10−4 are located within a 30-kb region and show high intermarker linkage disequilibrium (Fig. 1b). Two genes are located in close vicinity to this region: KIAA1598 (40 kb centromeric) and VAX1 (ventral anterior homeobox 1; 53 kb telomeric). Mice with homozygous Vax1 mutations display craniofacial malformations including cleft palate6. Two individuals with a 10q terminal deletion syndrome with breakpoints in 10q25 have been reported, one with a submucous cleft palate7 and the other with a cleft lip8.
Three further loci were successfully replicated but did not achieve genome-wide significance in the combined analysis. FMN1 (formin-1), a gene with an unknown function, and GREM1 (gremlin-1), the gene coding for another known antagonist of BMP4, are located on chromosome 15q13.3 (Supplementary Fig. 3a). The 13q31.1 locus lies in a gene desert (Supplementary Fig. 3b). SPRY2 (sprouty homolog 2) is located 241 kb telomeric of rs9574565. In mice, palate development is sensitive to Spry2 dosage9, and Spry2 overexpression results in craniofacial defects10. Resequencing of SPRY2 in NSCL/P cases has suggested the presence of rare, and possibly detrimental, variants in SPRY2 (ref. 11). The SNP on 2p21 (rs7590268) is located within intron 31 of THADA (thyroid adenoma associated), which may be involved in the cell death receptor pathway and apoptosis12 (Supplementary Fig. 3c). Notably, rs7590268 maps to a region that was duplicated in two individuals presenting with cleft palate and other anomalies13,14.
The most significant SNP at the 8q24.21 locus1 (rs987525) and the functional IRF6 variant2 (rs642961) were also genotyped in the replication sample in order to test for a possible interaction. Suggestive evidence (P = 0.005) was found for an interaction between the IRF6 variant rs642961 and the SNP near GREM1 at 15q13.3 (rs1258763). No evidence was found for any other interactions.
Recent genetic data2 support the hypothesis that NSCL/P may be separable into two sub-phenotypes: cleft lip only (NSCLO) and cleft lip with cleft palate (NSCLP). The genotype distribution in the NSCLP subsample (318 cases) did not differ significantly from that in the NSCLO subsample (81 cases) (data not shown) for any of the nine reported SNPs at the five newly identified loci (Supplementary Table 3).
Because NSCL/P and nonsyndromic cleft palate only (NSCPO) may have an etiological overlap, we genotyped the replication marker panel in 295 NSCPO triads. None of the SNPs showed evidence of association (Supplementary Table 4).
In summary, we have identified two new NSCL/P susceptibility loci with genome-wide significance on 17q22 and 10q25.3, and three further loci (13q31.1, 15q13.3 and 2p21) for which there is suggestive evidence. Promising candidate genes at these loci include NOG (noggin), VAX1 (ventral anterior homeobox 1), GREM1 (gremlin 1), SPRY2 (sprouty homolog 2) and THADA (thyroid adenoma associated). Given the intergenic location of the associated SNPs in the present study, further studies should test for allele-specific expression of these candidate genes and resequence their coding regions in order to identify possible functional variants.
Accession numbers. GenBank: BMP4, NM_001202 (NM_130850, NM_130851); FMN1, NM_001103184; GREM1, NM_013372; IRF6, NM_006147; KIAA1598, NM_018330 (NM_001127211); NOG, NM_005450; SPRY2, NM_005842; THADA, NM_022065; VAX1, NM_199131 (NM_001112704).
Accession codes
Accessions
NCBI Reference Sequence
References
Birnbaum, S. et al. Nat. Genet. 41, 473–477 (2009).
Rahimov, F. et al. Nat. Genet. 40, 1341–1347 (2008).
Guo, C.Y. et al. Genet. Epidemiol. 33, 54–62 (2009).
Zhang, Z. et al. Development 129, 4135–4146 (2002).
Suzuki, S. et al. Am. J. Hum. Genet. 84, 406–411 (2009).
Hallonet, M. et al. Genes Dev. 13, 3106–3114 (1999).
Petersen, B. et al. Am. J. Med. Genet. 77, 60–62 (1998).
Mulcahy, M.T. et al. Clin. Genet. 21, 33–35 (1982).
Welsh, I.C. et al. Mech. Dev. 124, 746–761 (2007).
Goodnough, L.H. et al. Dev. Dyn. 236, 1918–1928 (2007).
Vieira, A.R. et al. PLoS Genet. 1, e64 (2005).
Drieschner, N. et al. Gene 403, 110–117 (2007).
Yunis, E. et al. Hum. Genet. 48, 241–244 (1979).
Magee, A.C. et al. Clin. Genet. 54, 65–69 (1998).
Acknowledgements
We thank all affected individuals and their families for their participation in this study as well as the German support group for people with cleft lip and/or palate (Deutsche Selbsthilfevereinigung für Lippen-Gaumen-Fehlbildungen e.V.). We acknowledge our collaborating clinical partners, all colleagues who contributed to the European Collaboration on Craniofacial Anomalies (EUROCRAN) project, our laboratory technicians and colleagues responsible for database management (for a complete list of all individuals see Supplementary Note). The study was supported by the Deutsche Forschungsgemeinschaft (FOR 423 and individual grants MA 2546/3-1, KR 1912/7-1, NO 246/6-1 and WI 1555/5-1). The Heinz Nixdorf Recall cohort was established with the generous support of the Heinz Nixdorf Foundation, Germany (Chairman: G. Schmidt). POPGEN biobank is financed by the local Ministry of Science, Economy and Transport of Schleswig-Holstein, Germany. The Kooperative Gesundheitforschung in der Region Augsburg (KORA) research platform was initiated and financed by the Helmholtz Center Munich, German Research Center for Environmental Health, which is funded by the German Federal Ministry of Education and Research and by the state of Bavaria. The work of KORA is supported by the German Federal Ministry of Education and Research (BMBF) in the context of the German National Genome Research Network (NGFN-2 and NGFN-plus). The EUROCRAN study was supported by the European Commission FP5 (contract no. QLG1-CT-2000-01019), and the ITALCLEFT study was supported by a FAR-2008 grant from the University of Ferrara, Italy. N.A.d.A. is supported by the Conselho Nacional de Pesquisa (CNPq), Brasil. T.A.C. is supported by a grant from the Ministry of Higher Education, Syrian Arab Republic. Additional acknowledgments are found in the Supplementary Note.
Author information
Authors and Affiliations
Contributions
E.M., F.-J.K., T.F.W., P.P. and M.M.N. initiated the study. E.M., S. Birnbaum, K.U.L., P.H., M. Knapp, M.R., P.A.M. and M.M.N. contributed to the study design. M.M.N., E.M., S.C., P.H., K.U.L. and S. Birnbaum coordinated the work and prepared the manuscript, with feedback from the other authors. S. Birnbaum., H.R., A.P., C.L., G.S., M. Scheer., B. Braumann, R.H.R., A.H., S.P., B. Blaumeiser, R.P.S.-T., F.-J.K., M.R. and P.A.M. clinically characterized the cleft families and collected blood samples. S.M., M. Krawczak, S.S., T.M. and E.M. characterized and recruited the controls. K.U.L., C.B., M.F., N.A.d.A., T.A.C., S. Barth, N.K. and J.B. prepared the DNA and performed the molecular genetic experiments. M. Knapp., S.H., M. Steffens and M.M. conducted the statistical analysis. M.M.N., E.M., M. Knapp., T.F.W., S. Birnbaum, K.U.L., M.R. and P.P. analyzed and interpreted the data.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3, Supplementary Tables 1–4, Supplementary Methods and Supplementary Note (PDF 602 kb)
Rights and permissions
About this article
Cite this article
Mangold, E., Ludwig, K., Birnbaum, S. et al. Genome-wide association study identifies two susceptibility loci for nonsyndromic cleft lip with or without cleft palate. Nat Genet 42, 24–26 (2010). https://doi.org/10.1038/ng.506
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.506
This article is cited by
-
Genetic association and functional validation of ZFP36L2 in non-syndromic orofacial cleft subtypes
Journal of Human Genetics (2024)
-
Integrative analysis of transcriptome dynamics during human craniofacial development identifies candidate disease genes
Nature Communications (2023)
-
Global perspectives in orofacial cleft management and research
British Dental Journal (2023)
-
Using whole exome sequencing to identify susceptibility genes associated with nonsyndromic cleft lip with or without cleft palate
Molecular Genetics and Genomics (2023)
-
Rare variant modifier analysis identifies variants in SEC24D associated with orofacial cleft subtypes
Human Genetics (2023)