Abstract
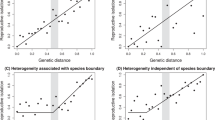
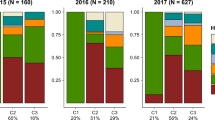
Parallel evolution of similar traits in independent populations that inhabit ecologically similar environments strongly implicates natural selection as the cause of evolution1. Parallel speciation is a special form of parallel evolution where traits that determine reproductive isolation evolve repeatedly, in closely related populations, as by-products of adaptation to ecological conditions1,2. The outcome of such parallel evolution is that ecologically divergent pairs of populations exhibit greater levels of reproductive isolation than ecologically similar pairs of populations of a similar or younger age2,3,4. The parallel evolution of reproductive isolation provides strong evidence for natural selection in the process of speciation1, but only one conclusive example from nature is known2. Populations of the walking-stick insect Timema cristinae that use different host-plant species have diverged in body size and shape, host preference, behaviour and the relative frequency of two highly cryptic colour-pattern morphs5,6. Here we report that divergent selection for host adaptation, and not genetic drift, has promoted the parallel evolution of sexual isolation in this species. Our findings represent a clear demonstration that host-plant adaptation can play a crucial and repeatable role in the early stages of speciation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Schluter, D. & Nagel, L. Parallel speciation by natural selection. Am. Nat. 146, 292–301 (1995)
Rundle, H. D., Nagel, L., Boughman, J. W. & Schluter, D. Natural selection and parallel speciation in sticklebacks. Science 287, 306–308 (2000)
Funk, D. J. Isolating a role for natural selection in speciation: host adaptation and sexual isolation in Neochlamisus bebbianae leaf beetles. Evolution 52, 1744–1759 (1998)
McPeek, M. A. & Wellborn, G. A. Genetic variation and reproductive isolation among phenotypically divergent amphipod populations. Limnol. Oceonogr. 43, 1162–1169 (1998)
Sandoval, C. P. Differential visual predation on morphs of Timema cristinae (Phasmatodeae: Timemidae) and its consequences for host range. Biol. J. Linn. Soc. 52, 341–356 (1994)
Sandoval, C. P. The effects of relative geographic scales of gene flow and selection on morph frequencies in the walking stick Timema cristinae. Evolution 48, 1866–1879 (1994)
Crespi, B. J. & Sandoval, C. P. Phylogenetic evidence for the evolution of ecological specialization in Timema walking-sticks. J. Evol. Biol. 13, 249–262 (2000)
Sandoval, C. P. Ecological and Behavioral Factors Affecting Spatial Variation in Colour or Morph Frequency in the Walking-stick Timema cristinae. Thesis, Univ. California, Santa Barbara (1993)
Shimodaira, H. & Hasegawa, M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 16, 1114–1116 (1999)
Templeton, A. R. Phylogenetic inference from restriction endonuclease cleavage site maps with particular reference to the evolution of humans and apes. Evolution 37, 221–244 (1983)
Manly, B. F. J. Randomization, Bootstrap and Monte Carlo Methods in Biology (Chapman and Hall, London, 1997)
Rolàn-Alvarez, E. & Caballero, A. Estimating sexual selection and sexual isolation effects from mating frequencies. Evolution 54, 30–36 (2000)
Simon, C. et al. Evolution, weighting and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann. Ent. Soc. Am. 87, 651–701 (1994)
Posada, D. & Crandall, K. A. MODELTEST: testing the model of DNA substitution. Bioinformatics 14, 817–818 (1998)
Swofford, D. L. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods) 4.0b4a (Sinauer Associates, Sunderland, Massachusetts, 2001)
Huelsenbeck, J. P. & Crandall, K. A. Phylogeny estimation and hypothesis testing using maximum likelihood. Annu. Rev. Ecol. Syst. 28, 437–466 (1997)
Acknowledgements
We thank T. E. Reimchen, H. Rundle, A. Mooers, F. Breden, J. Endler, R. Vos, C. Parent, J. Joy, S. Springer and especially D. Schluter for discussion and comments on the manuscript. M. Fulton, D. McLaren, B. Mickelson, M. Vankoeveringe and T. Luchin provided field and technical assistance. E. Rolàn-Alvarez provided software and statistical advice. J. Endler provided laboratory space for all the mating experiments. Financial support was provided by the Natural Science and Engineering Research Council of Canada.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Rights and permissions
About this article
Cite this article
Nosil, P., Crespi, B. & Sandoval, C. Host-plant adaptation drives the parallel evolution of reproductive isolation. Nature 417, 440–443 (2002). https://doi.org/10.1038/417440a
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/417440a
This article is cited by
-
Hybridisation between host races broadens the host range of offspring in Eotetranychus asiaticus (Acari: Tetranychidae)
Experimental and Applied Acarology (2023)
-
Host-plant adaptation as a driver of incipient speciation in the fall armyworm (Spodoptera frugiperda)
BMC Ecology and Evolution (2022)
-
Non-parallel morphological divergence following colonization of a new host plant
Evolutionary Ecology (2022)
-
Insight into incipient reproductive isolation in diverging populations of Brachionus plicatilis rotifer
Hydrobiologia (2022)
-
What are fungal species and how to delineate them?
Fungal Diversity (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.