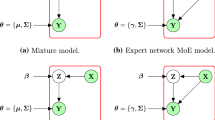
Abstract
Covariate informed product partition models incorporate the intuitively appealing notion that individuals or units with similar covariate values a priori have a higher probability of co-clustering than those with dissimilar covariate values. These methods have been shown to perform well if the number of covariates is relatively small. However, as the number of covariates increase, their influence on partition probabilities overwhelm any information the response may provide in clustering and often encourage partitions with either a large number of singleton clusters or one large cluster resulting in poor model fit and poor out-of-sample prediction. This same phenomenon is observed in Bayesian nonparametric regression methods that induce a conditional distribution for the response given covariates through a joint model. In light of this, we propose two methods that calibrate the covariate-dependent partition model by capping the influence that covariates have on partition probabilities. We demonstrate the new methods’ utility using simulation and two publicly available datasets.









Similar content being viewed by others
References
Antoniano-Villalobos, I., Walker, S.G.: A nonparametric model for stationary time series. J. Time Ser. Anal. 37(1), 126–142 (2016)
Barcella, W., Iorio, M.D., Baio, G.: A comparative review of variable selection techniques for covariate dependent Dirichlet process mixture models (2016). https://arxiv.org/pdf/1508.00129.pdf
Barcella, W., Iorio, M.D., Baio, G., Malone-Lee, J.: Variable selection in covariate dependent random partition models: an application to urinary tract infection. Stat. Med. 35, 1373–1389 (2016)
Barrientos, A.F., Jara, A., Quintana, F.A.: On the support of MacEachern’s dependent Dirichlet processes and extensions. Bayes Anal. 7, 277–310 (2012)
Blei, D.M., Frazier, P.I.: Distant dependent chinese restaurant processes. J. Mach. Learn. Res. 12, 2461–2488 (2011)
Christensen, R., Johnson, W., Branscum, A.J., Hanson, T.: Bayesian Ideas and Data Analysis: An Introduction for Scientists and Statisticians. CRC Press, Boca Raton (2011). http://www.ics.uci.edu/~wjohnson/BIDA/BIDABook.html
Chung, Y., Dunson, D.B.: Nonparametric bayes conditional distribution modeling with variable selection. J. Am. Stat. Assoc. 104, 1646–1660 (2009)
Cook, R.D., Weisberg, S.: Sliced inverse regression for dimension reduction: comment. J. Am. Stat. Assoc. 86, 328–332 (1991)
Dahl, D.B.: Model-based clustering for expression data via a Dirichlet process mixture model. In: Vannucci, M., Do, K.A., Müller, P. (eds.) Bayesian Inference for Gene Expression and Proteomics, pp. 201–218. Cambridge University Press, Cambridge (2006)
Dahl, D.B., Day, R., Tsai, J.W.: Random partition distribution indexed by pairwise information. J. Am. Stat. Assoc. (2016). doi:10.1080/01621459.2016.1165103
De Iorio, M., Müller, P., Rosner, G., MacEachern, S.: An ANOVA model for dependent random measures. J. Am. Stat. Assoc. 99, 205–215 (2004)
Dunson, D.B., Park, J.H.: Kernel stick-breaking processes. Biometrika 95, 307–323 (2008)
Geisser, S., Eddy, W.F.: A predictive approach to model selection. J. Am. Stat. Assoc. 74(365), 153–160 (1979)
Gelfand, A.E., Kottas, A., MacEachern, S.N.: Bayesian nonparametric spatial modeling with Dirichlet process mixing. J. Am. Stat. Assoc. 102, 1021–1035 (2005)
Gower, J.C.: A general coefficient of similarity and some of its properties. Biometrics 27, 857–871 (1971)
Griffin, J.E., Steel, M.F.J.: Order-based dependent Dirichlet processes. J. Am. Stat. Assoc. 101, 179–194 (2006)
Guhaniyogi, R., Dunson, D.B.: Bayesian compressed regression. J. Am. Stat. Assoc. 110, 1500–1514 (2015)
Hannah, L., Blei, D., Powell, W.: Dirichlet process mixtures of generalized linear models. J. Mach. Learn. Res. 12, 1923–1953 (2011)
Hartigan, J.A.: Partition models. Commun. Stat. Theory Methods 19, 2745–2756 (1990)
Jacobs, R.A., Jordan, M.I., Nowlan, S.J., Hinton, G.E.: Adaptive mixtures of local experts. Neural Comput. 3, 79–87 (1991)
Lichman, M.: UCI machine learning repository (2013). http://archive.ics.uci.edu/ml
MacEachern, S.N.: Dependent Dirichlet processes. Ohio State University, Department of Statistics, Technical report (2000)
Maechler, M., Rousseeuw, P., Struyf, A., Hubert, M., Hornik, K.: Cluster: Cluster Analysis Basics and Extensions (2016). R package version 2.0.4—For new features, see the ’Changelog’ file (in the package source)
McLachlan, G., Peel, D.: Finite Mixture Models, 1st edn. Wiley Series in Probability and Statistics, New York (2000)
Miller, J.W., Dunson, D.B.: Robust Bayesian inference via coarsening (2015). http://arxiv.org/abs/arXiv:1506.06101
Molitor, J., Papathomas, M., Jerrett, M., Richardson, S.: Random partition models with regression on covariates. Biostatistics 11, 484–498 (2010)
Müller, P., Erkanli, A., West, M.: Bayesian curve fitting using multivariate normal mixutres. Biometrika 83, 67–79 (1996)
Müller, P., Quintana, F.A., Jara, A., Hanson, T.: Bayesian Nonparametric Data Analysis, 1st edn. Springer, Switzerland (2015)
Müller, P., Quintana, F.A., Rosner, G.L.: A product partition model with regression on covariates. J. Comput. Graph. Stat. 20(1), 260–277 (2011)
Müller, P., Quintana, F.A., Rosner, G.L., Maitland, M.L.: Bayesian inference for longitudinal data with non-parametric treatment effects. Biostatistics 15(2), 341–352 (2013)
Neal, R.M.: Markov chain sampling methods for Dirichlet process mixture models. J. Comput. Graph. Stat. 9, 249–265 (2000)
Page, G.L., Bhattacharya, A., Dunson, D.B.: Classification via Bayesian nonparametric learning of affine subspaces. J. Am. Stat. Assoc. 108, 187–201 (2013)
Page, G.L., Quintana, F.A.: Predictions based on the clustering of heterogeneous functions via shape and subject-specific covariates. Bayesian Anal. 10, 379–410 (2015)
Page, G.L., Quintana, F.A.: Spatial product partition models. Bayesian Anal. 11(1), 265–298 (2016)
Papathomas, M., Molitor, J., Hoggart, C., Hastie, D., Richardson, S.: Exploring data from genetic association studies using bayesian variable selection and the Dirichlet process: application to searchingfor gene \(\times \) gene patterns. Genet. Epidemiol. 36, 663–674 (2012)
Park, J.H., Dunson, D.B.: Bayesian generalized product partition model. Stat. Sin. 20, 1203–1226 (2010)
Quintana, F.A., Müller, P., Papoila, A.L.: Cluster-specific variable selection for product partition models. Scand. J. Stat. 42, 1065–1077 (2015). doi:10.1111/sjos.12151
R Core Team: R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria (2016). https://www.R-project.org/
Rand, W.M.: Objective criteria for the evaluation of clustering methods. J. Am. Stat. Assoc. 66, 846–850 (1971)
Rodriguez, A., Dunson, D.B., Gelfand, A.E.: Bayesian nonparametric functional data analysis through density estimation. Biometrika 96, 149–162 (2009)
Wade, S., Dunson, D.B., Petrone, S., Trippa, L.: Improving prediction from Dirichlet process mixtures via enrichment. J. Mach. Learn. Res. 15, 1041–1071 (2014)
Wang, H., Xia, Y.: Sliced regression for dimension reduction. J. Am. Stat. Assoc. 103, 811–821 (2008)
Acknowledgements
The authors would like to thank Peter Müller for helpful comments. The authors also thank all the reviewers for their valuable suggestions that substantially improved presentation. Garritt L. Page gratefully acknowledges the financial support of FONDECYT Grant 11121131 and Fernando A. Quintana was partially funded by Grant FONDECYT 1141057.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Appendices
Appendix: MCMC algorithm for the calibrated similarity and tempered mixture of experts
Here we provide pertinent computation details for the MCMC algorithm used to fit the TME model and PPMx with calibrated similarity. We focus primarily on the updating of cluster labels, as conditional on these updating the remaining model parameters is straightforward employing a Gibbs sampler or Metropolis–Hastings steps.
1.1 Calibrated similarity
To update the cluster membership of subject i for the calibrated similarity, cluster weights are created by comparing the unnormalized posterior for the jth cluster when subject i is excluded from that when subject i is included. In addition to weights for existing clusters, algorithm 8 of Neal (2000) requires calculating weights for p empty clusters whose cluster-specific parameters are auxiliary variables generated from the prior. To make this more concrete, let \(S_j^{-i}\) denote the jth cluster and \(k^{-i}\) the number of clusters when subject i is not considered. Similarly \(\varvec{x}_j^{\star -i}\) will denote the vector of covariates corresponding to cluster h when subject i has been removed. Then the multinomial weights associated with the \(k^{-i}\) existing clusters and one empty cluster are
where as mentioned \(\mu ^{\star }_{\mathrm{new}, j}\) and \(\sigma ^{2\star }_{\mathrm{new},j}\) are auxiliary variables that are drawn from their respective prior distributions. Since \(\tilde{g}(\{\varvec{x}_i\})\) needs to be accounted for when standardizing the multinomial weights, we employ the following ratios in the MCMC algorithm
When \(\varvec{x}_i\) is included in the j cluster then it is not able to form its own singleton. However, when it is excluded from the jth cluster, then it is completely plausible that it forms its own singleton cluster. For these reasons the similarity value \(g(\{\varvec{x}_i\})\) is only included in \(\tilde{g}(\varvec{x}^{\star -i}_{j})\) and \(\tilde{g}(\{\varvec{x}_i\})\).
1.2 TME with unknown \(\varvec{\xi }^{\star }_j\) and fixed J
Upon introducing latent component labels \(s_i\) such that \(Pr(s_i = j) = w(\varvec{x}_i; \varvec{\xi }^{\star }_j)\), the data model (14) can be written hierarchically as
where \(\delta _{\ell }\) is the dirac measure. With this hierarchical representation, a MCMC algorithm can be constructed by cycling through the following
-
Update component labels using
$$\begin{aligned} Pr(s_i = h | - )&\propto N(y_i | \mu ^{\star }_h, \sigma ^{2\star }_h) w(\varvec{x}_i ; \varvec{\xi }^{\star }_h) \end{aligned}$$ -
If \(\varvec{x}_i\) is comprised of continuous and categorical variables, then without loss of generality let \(\varvec{x}_i = (x_{1i}, x_{2i})\) where \(x_{1i}\) is continuous and \(x_{2i}\) is categorical. Further, \(\varvec{\xi }_j^{\star } = (\eta ^{\star }_j,v^{2\star }_j, \varvec{\pi }^{\star }_j)\) with \(\varvec{\xi }^{\star } = (\varvec{\xi }_1^{\star },\ldots , \varvec{\xi }_J^{\star })\). Then \(\varvec{\xi }_j^{\star }=(\eta ^{\star }_j,v^{2\star }_j, \varvec{\pi }^{\star }_j)\) can be updated within the MCMC algorithm by way of a Metroplis–Hastings step employing
$$\begin{aligned} {[}\varvec{\xi }^{\star }_j | - ]&\propto \prod _{i=1}^m Pr(s_i | \varvec{\xi }^{\star })\prod _{j=1}^J p(\varvec{\xi }^{\star }_j) \\&\propto \prod _{i=1}^m w(\varvec{x}_{i}; \varvec{\xi }^{\star }_1)^{I[s_i=1]} \times \ldots \times w(\varvec{x}_{i}; \varvec{\xi }^{\star }_1)^{I[s_i=J]} p(\varvec{\xi }^{\star }_j) \\&\propto \prod _{i:s_i=j} w(\varvec{x}_{i}; \varvec{\xi }^{\star }_j) p(\varvec{\xi }^{\star }_j)\\&= \prod _{i:s_i=j} \frac{q(x_{i1}|\eta ^{\star }_j, v^{2\star }_j) q(x_{i2}|\varvec{\pi }^{\star }_j)}{\sum _{\ell }^Jq (x_{i1}|\eta ^{\star }_{\ell }, v^{2\star }_{\ell })q(x_{i2}|\varvec{\pi }^{\star }_{\ell })}p(\eta ^{\star }_j,v^{2\star }_j, \varvec{\pi }^{\star }_j) \end{aligned}$$where \(q(x_{i1}|\eta ^{\star }_j, v^{2\star }_j)\) is normal density and and \(q(x_{i2}|\varvec{\pi }^{\star }_j)\) a multinomial density. For \(\varvec{\pi }^{\star }_j\) an independent Metropolis–Hastings sampler with uniform (over the simplex) candidate density may be considered. This candidate density will cancel out in the Metropolis–Hastings ratio (though this may become more inefficient as the number of categories in \(x_{i2}\) increases). Updating \(\eta ^{\star }_j\) and \(v^{2\star }_j\) can be accomplished using a random walk Metropolis step with normal candidate density for both.
-
Updating the likelihood parameters \(\mu _j^{\star }\) and \(\sigma _j^{2\star }\) can be carried out using Gibbs steps as their full conditionals have well known closed forms.
Computing Gower’s dissimilarity
The daisy function found in the cluster package (Maechler et al. 2016) of the statistical software R was employed to calculate the Gower dissimilarity. The calculated dissimilarity is an “average” of the individual p dissimilarities
For numeric or continuous x’s, \(d(x_{i\ell },x_{j\ell }) = |x_{i\ell } - x_{j\ell }|/R_{\ell }\) where \(R_{\ell } = \max _h(x_{h\ell }) - \min _h(x_{h\ell })\). For nominal variables
Rights and permissions
About this article
Cite this article
Page, G.L., Quintana, F.A. Calibrating covariate informed product partition models. Stat Comput 28, 1009–1031 (2018). https://doi.org/10.1007/s11222-017-9777-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11222-017-9777-z