Novel targets for engineering Physostegia chlorotic mottle and tomato brown rugose fruit virus-resistant tomatoes: in silico prediction of tomato microRNA targets
- Published
- Accepted
- Received
- Academic Editor
- Savithramma Dinesh-Kumar
- Subject Areas
- Bioinformatics, Computational Biology, Genomics, Plant Science, Virology
- Keywords
- miRNA, Alphanucleorhabdovirus, Tobamovirus, PhCMoV, ToBRFV, Resistance, Transgenes, RNA interference, Solanum lycopersicum
- Copyright
- © 2020 Gaafar and Ziebell
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits using, remixing, and building upon the work non-commercially, as long as it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2020. Novel targets for engineering Physostegia chlorotic mottle and tomato brown rugose fruit virus-resistant tomatoes: in silico prediction of tomato microRNA targets. PeerJ 8:e10096 https://doi.org/10.7717/peerj.10096
Abstract
Background
Physostegia chlorotic mottle virus (PhCMoV; genus: Alphanucleorhabdovirus, family: Rhabdoviridae) and tomato brown rugose fruit virus (ToBRFV; genus: Tobamovirus, family: Virgaviridae) are newly emerging plant viruses that have a dramatic effect on tomato production. Among various known virus-control strategies, RNAi-mediated defence has shown the potential to protect plants against various pathogens including viral infections. Micro(mi)RNAs play a major role in RNAi-mediated defence.
Methods
Using in silico analyses, we investigated the possibility of tomato-encoded miRNAs (TomiRNA) to target PhCMoV and ToBRFV genomes using five different algorithms, i.e., miRanda, RNAhybrid, RNA22, Tapirhybrid and psRNATarget.
Results
The results revealed that 14 loci on PhCMoV and 10 loci on ToBRFV can be targeted by the TomiRNAs based on the prediction of at least three algorithms. Interestingly, one TomiRNA, miR6026, can target open reading frames from both viruses, i.e., the phosphoprotein encoding gene of PhCMoV, and the two replicase components of ToBRFV. There are currently no commercially available PhCMoV- or ToBRFV-resistant tomato varieties, therefore the predicted data provide useful information for the development of PhCMoV- and ToBFRV-resistant tomato plants.
Introduction
Tomato (Solanum lycopersicum) is an economically important vegetable crop for human consumption as a fresh crop and as an ingredient in many prepared foods; it is also used as a model in fundamental research areas such as plant growth and fruit development (Hobson & Grierson, 2012). The production of tomato continues increasing worldwide. They are grown as annuals and as facultative perennial plants (Rick, 1974). The tomato genome possesses a haploid set of 12 chromosomes, and the genome of tomato was sequenced in 2012 (The Tomato Genome Consortium, 2012).
Tomato production is hampered by different pathogens including fungi, nematodes and viruses (Moriones & Navas-Castillo, 2000; Kiss et al., 2001; Zia et al., 2014). Several viruses from different families affect tomatoes worldwide and are responsible for serious yield losses. Viruses can cause a wide range of symptoms including marbling of fruits, leaf distortion and deformation, mosaic and stunting. Important tomato viruses include tomato yellow leaf curl virus (TYLCV; genus: Begomovirus, family: Geminiviridae), pepino mosaic virus (PepMV; genus Potexvirus, family Alphaflexiviridae), tobamoviruses (family: Virgaviridae), e.g., tobacco mosaic virus (TMV) and tomato mosaic virus (ToMV), tomato spotted wilt virus (TSWV; genus: Tospovirus, family: Peribunyaviridae), tomato torrado virus (ToTV; genus: Torradovirus, family: Secoviridae), criniviruses (family: Closteroviridae) i.e., tomato chlorosis virus (ToCV) and tomato infectious chlorosis virus (TICV) and cucumber mosaic virus (CMV; genus: Cucumovirus, family: Bromoviridae), amongst others (Best, 1968; Pelham, 1972; Broadbent, 1976; Jordá, 1992; Duffus, Liu & Wisler, 1996; Wisler et al., 1998; Moriones & Navas-Castillo, 2000; Verbeek et al., 2007; Hanssen & Thomma, 2010). Tomato viruses are transmitted by different means such as fungi, insects (by aphids, thrips, whiteflies, leafhoppers and treehoppers), or mechanically by tools or human handling of crops whereas seed transmission is also possible for some viruses (Sakimura, 1962; Amari et al., 2008; Alfaro-Fernandez, 2010; Hanssen et al., 2010; Jeger et al., 2017; Ong et al., 2020).
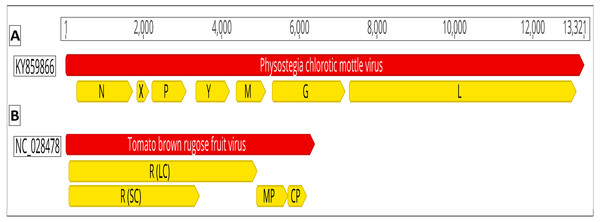
Recently, two new viruses were discovered that cause severe symptoms on tomato plants i.e., Physostegia chlorotic mottle virus (PhCMoV; genus: Alphanucleorhabdovirus, family: Rhabdoviridae) and tomato brown rugose fruit virus (ToBRFV; a tobamovirus) (Salem et al., 2016; Gaafar et al., 2018). PhCMoV was first detected in Physostegia virginiana from Austria (Menzel et al., 2016). PhCMoV was found to infect tomatoes in Germany causing severe fruit marbling (Gaafar et al., 2018). The virions of PhCMoV are bacilliform containing (-ve) ssRNAs. PhCMoV’s genome consists of seven open reading frames (ORF) which are predicted to encode the nucleocapsid [N], phospho- [P], movement [Y], matrix [M], glyco- [G], RNA dependent RNA polymerase/large [L] proteins, and the X protein (with unknown function) (Fig. 1). Although PhCMoV can be transmitted mechanically, its natural dispersal pathways are currently unknown.
Figure 1: Genomic organization of (A) Physostegia chlorotic mottle virus (PhCMoV; KY859866) and (B) tomato brown rugose fruit virus (ToBRFV; NC_028478).
The open reading frames are shown in yellow. PhCMoV; N, nucleocapsid, X, hypothetical protein with unknown function; P, phosphoprotein; Y, movement protein; M, matrix protein; G, glycoprotein and L, Large protein/RNA dependent RNA polymerase. ToBRFV; R (LC), replicase large component “subunit”; R (SC), replicase small component; MP, movement protein and CP, capsid protein.ToBRFV was reported from several countries in the Middle East, Europe, America and China (Salem et al., 2016; Alkowni, Alabdallah & Fadda, 2019; Ling et al., 2019; Menzel et al., 2019; Panno, Caruso & Davino, 2019; Yan et al., 2019). The virions of ToBRFV are rod-shaped, and their genome consists of (+ve) ssRNA with four ORFs that encode the large (LC) and the small (SC) replicase components “subunits” as well as the movement (MP) and the capsid (CP) proteins (Fig. 1).
Plant viruses are unique amongst plant diseases in a way that once an infection has taken place, no cure is available. The control of viral vectors, e.g., insects or fungi, by pesticides is often not effective to control the virus disease. Additionally, many viruses are transmitted mechanically, thus requiring strict hygiene measures to prevent virus outbreaks. Although the use of virus-resistant varieties is the preferred and most successful way to prevent virus induced crop losses, not many commercial virus-resistant tomato varieties are available. Resistance is limited to a few numbers of viruses including TMV, ToMV, TSWV and TYLCV (Reimer Seeds, 2020a; Reimer Seeds, 2020b; Reimer Seeds, 2020c). Currently, there are no PhCMoV- and ToBRFV-resistant tomato varieties available.
Plant microRNAs (miRNAs) are endogenous non-coding small RNAs of 21 to 24 nucleotides in length (Jin et al., 2013). Their precursor RNAs have hairpin-like secondary structures (Starega-Roslan et al., 2011). miRNA precursors are processed by Dicer-like (DCL) enzymes and converted into mature miRNAs (mat-miRNAs) (Wang et al., 2018b). The mat-miRNAs join the RNA-induced silencing complex (RISC) which binds and suppresses the target transcripts at transcriptional or post-transcriptional levels (Baulcombe, 2004; Rogers & Chen, 2013; Borges & Martienssen, 2015). Together with small interfering RNAs (siRNAs), they are part of the plant small RNAs (sRNAs) that are involved in the cytoplasmic pathways of RNA silencing (Fang & Qi, 2016; Wang et al., 2018b). Thus, they can regulate the growth, development, genome stability and response of plants to both biotic and abiotic stresses (Jin et al., 2013).
Transgenic plants expressing artificial microRNAs (amiRNAs) have successfully been used to provide tolerance or resistance to virus infections caused by begomo-, cucumo- and orthotospoviruses (Zhang et al., 2011; Ali et al., 2013; van Vu, Choudhury & Mukherjee, 2013; Mitter et al., 2016). For example, transgenic tomato plants expressing amiRNAs targeting the transcripts of the pre-coat and coat proteins encoding sequences of tomato leaf curl New Delhi virus (ToLCNDV) showed tolerance to the virus infection (van Vu, Choudhury & Mukherjee, 2013). Moreover, transgenic tomato plants expressing amiRNAs, targeting the 2a and 2b genes and the 3′ untranslated conserved region of CMV, displayed effective resistance to CMV infection, and CMV in mixed infections with non-targeted viruses, including TMV and TYLCV (Zhang et al., 2011).
Plant miRNA-mRNA binding depends on a high quality match between the target sequence and the miRNA (Witkos, Koscianska & Krzyzosiak, 2011). Computational tools are available that predict several miRNA target sites within virus sequences (Pradhan et al., 2015; Iqbal et al., 2016; Iqbal et al., 2017; Jabbar et al., 2019). To study the possible interactions between tomato miRNAs (TomiRNA) and the PhCMoV and ToBRFV genomes, we used five different bioinformatic algorithms to predict the TomiRNA binding to the PhCMoV and ToBRFV genome sequences. Their analyses provide useful information to aid the development of PhCMoV- and ToBRFV-resistant tomato plants using amiRNA.
Materials & Methods
Sources and data retrieving
The available full genome sequences of different Physostegia chlorotic mottle virus isolates (PhCMoV; accession no. KX636164, KY706238, KY859866 and MK948541) as well as tomato brown rugose fruit virus isolates (ToBRFV NC_028478, KX619418, MN013187, MN013188, MK133095, MK165457, MN167466, MK133093, MK648157, MN182533 and MK319944) were obtained from NCBI GenBank. A total of 147 tomato (S. lycopersicum; sly) mature miRNA sequences (TomiRNA; commonly called sly-miRNA) were obtained from miRBase website (http://www.mirbase.org/) and used for this study (Table S1).
In silico microRNA target prediction analyses
For each virus, the retrieved sequences were aligned using MUSCLE tool [maxiters=16] (Edgar, 2004; Cuccuru et al., 2014) on Galaxy server (https://usegalaxy.eu/). The generated consensus sequences were visualized on Geneious Prime (2020.1.2) and open reading frames were predicted using Find ORFs tool. The generated consensus sequences of both viruses were used for further analyses. To predict the TomiRNA target sites, five different bioinformatic tools were used, i.e., miRanda, RNAhybrid, RNA22, Tapirhybrid and psRNATarget (Enright et al., 2003; Krüger & Rehmsmeier, 2006; Miranda et al., 2006; Bonnet et al., 2010; Srivastava et al., 2014; Dai, Zhuang & Zhao, 2018). The parameters used for each tool are shown in Table 1. The precursor of the potential microRNAs (pre-miRNA) were folded using RNAfold (Gruber et al., 2008; Lorenz et al., 2011).
| Tool | Parameters | Reference/source |
|---|---|---|
| miRanda | Score threshold = 140, energy threshold = −20 kcal/mol |
Enright et al. (2003) Run on Galaxy server: https://usegalaxy.eu/ |
| RNAhybrid | The E-value was set to −20 kcal/mol, and the remainder of the parameters were set to default |
Srivastava et al. (2014) https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid/ |
| RNA22 | Minimum number of paired-up bases was kept to 12 while the maximum folding energy was kept at −14 kcal/mol |
Miranda et al. (2006) https://cm.jefferson.edu/rna22/Interactive/ |
| Tapirhybrid | Score <= 8 and mfe_ratio >= 0.5 |
Bonnet et al. (2010) http://bioinformatics.psb.ugent.be/webtools/tapir/ |
| psRNATarget | Minimum expectation score = 7.0, extending gap = 0.5, opening gap = 2, G.U pair = 1, other mismatches = 1, HSP size=19, seed region = 2–7 nucleotides. |
Dai, Zhuang & Zhao (2018) http://plantgrn.noble.org/psRNATarget/analysis?function=3 |
Statistical analysis
TomiRNA predicted data obtained from all the five bioinformatic tools were analysed using scripts written on R statistical software (R Core Team, 2013). For graphical representation of the result, ggplot2 and limma packages were used (Ritchie et al., 2015; Wickham, 2016).
Results and Discussion
In the last years, two emerging viruses, i.e., PhCMoV and ToBRFV, have affected the production of tomatoes in Europe and worldwide, respectively. There are currently no known tomato varieties resistant to these viruses, thus alternative solutions are required, e.g., the production of transgenic plants.
Plant viruses can be targeted by host plant miRNAs (Chen, 2011). In recent years, endogenous miRNAs as important regulators of gene expression have also been used in functional genetic studies and for crops genetic improvement (Sablok et al., 2011). Moreover, amiRNA is used as a gene regulation strategy, designed to target e.g., pathogen genes. Transgenic plants producing amiRNA were shown to be resistant or tolerant to viral infection (Zhang et al., 2011; Ali et al., 2013). By using computational approaches, we can predict host miRNA targeting sites within the genome of the viruses, thus helping us to choose possible candidates prior to engineering or transformation (Xia, Cao & Shao, 2009; Witkos, Koscianska & Krzyzosiak, 2011; Peterson et al., 2014; Iqbal et al., 2017).
Various miRNA target prediction and identification algorithms have been investigated for their accuracy and efficiency (Bartel, 2009; Xia, Cao & Shao, 2009; Witkos, Koscianska & Krzyzosiak, 2011; Srivastava et al., 2014). We selected five different bioinformatic algorithms (miRanda, RNAhybrid, RNA22, Tapirhybrid and psRNATarget) for this study based on their performance and we used the recommended parameters for the folding energy, seed pairing, target site accessibility and pattern recognition, and ensuring the minimum free energy (MFE) exceeding the threshold standards (Enright et al., 2003; Krüger & Rehmsmeier, 2006; Miranda et al., 2006; Bonnet et al., 2010; Srivastava et al., 2014; Iqbal et al., 2016; Iqbal et al., 2017; Dai, Zhuang & Zhao, 2018; Jabbar et al., 2019). Therefore, the approach used here allowed a low number of mismatches in miRNA binding sites, to reduce most of the falsely predicted target loci.
These five algorithms were developed to identify small RNAs’ target loci by different approaches. miRanda considers for the prediction: the sequence complementarity, free energy of miRNA-target duplex and the cross-species conservation of the target site (John et al., 2004). It can also predict multiple target loci (John et al., 2004). RNAhybrid analyses the loci sequence complementarity, target-site abundance and the MFE (Krüger & Rehmsmeier, 2006). RNA22 identifies the target loci by implementing a different approach i.e., the pattern-based approach and the folding energy (Miranda et al., 2006). It does not rely on cross-species conservation (Miranda et al., 2006). Moreover, its algorithm analyses the target sequence for putative miRNA binding sites then defines the targeting miRNAs (Miranda et al., 2006). Tapirhybrid is a highly recommended plant miRNA target prediction tool due to its precise algorithm (Bonnet et al., 2010; Srivastava et al., 2014). It considers seed pairing, target site accessibility and multiple target sites (Bonnet et al., 2010). psRNATarget analyses complementary matching between the miRNA sequence and target sequence using a scoring schema and evaluates target site accessibility (Dai, Zhuang & Zhao, 2018). The analytical performance of psRNATarget is enhanced by the developing of its new scoring schema that is able to discover miRNA-target interactions at higher rates (Dai, Zhuang & Zhao, 2018).
The five algorithms identified possible target sites for TomiRNAs within the genomes of the two viruses. All TomiRNAs used in this study can target the genome of one or both viruses as predicted by at least one tool (Table S1).
TomiRNAs’ target loci on Physostegia chlorotic mottle virus genome:
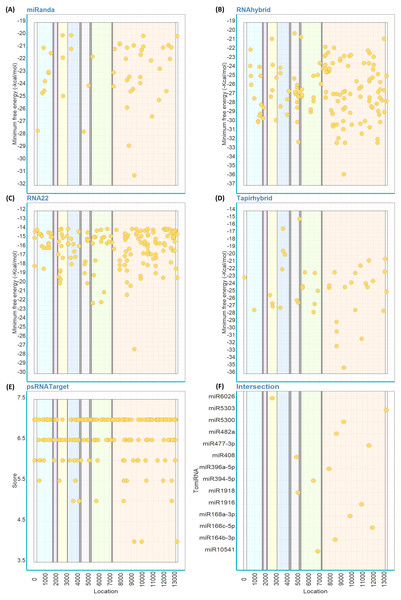
Out of the 147 mature TomiRNAs, miRanda predicted that 38 TomiRNAs target 49 loci on PhCMoV genome (Fig. 2A). RNAhybrid predicted that 145 TomiRNAs target 145 loci (Fig. 2B), RNA22: 74 TomiRNAs and 170 loci (Fig. 2C), Tapirhybrid: 41 TomiRNAs and 46 loci (Fig. 2D), and psRNATarget: 107 TomiRNAs and 226 loci (Fig. 2E). Table 2 shows the number of locations targeted by the TomiRNAs by the five different algorithms used in this study.
Figure 2: Predicted target sites of tomato’s miRNAs on the genome of PhCMoV using miRanda (A), RNAhybrid (B), RNA22 (C), Tapirhybrid (D), psRNATarget (E) and (F) shows the common loci predicted by at least three miRNA target prediction algorithms.
The genes are highlighted as follow; N (light blue), X (violet), P (yellow), Y (blue), M (grey), G (green) and L (orange), and the 3′and 5′-UTRs (white).Only 14 TomiRNAs have common loci on the PhCMoV genome as predicted by at least three algorithms (Fig. 2F). Four out of the seven predicted genes of PhCMoV, i.e., P, M, G and L, are targets by TomiRNAs as identified by at least three algorithms (Fig. 2F). For the genes N, X and Y, only two or less algorithms were able to predict miRNA targets. Eight TomiRNAs are targeting the L gene sequence, i.e., miR396a-5p (nucleotide [nt] start position 7925), miR164b-3p (8494), miR482a (8611), miR5300 (9285), miR168a-3p (9878), miR1916 (10935), miR477-3p (11628) and miR166c-5p (11956). Two TomiRNAs are targeting the M gene, i.e., miR408 at 4929 and miR1918 at 5058, and two are targeting the G gene i.e., miR394-5p at 6486 and miR10541 at 6874 (Fig. 2F). miR6026 is only targeting the P gene at nt position 2663 and miR5303 is targeting the 5′ end at nt position 13248 (Fig. 2F). The predicted folding structures of the precursor miRNAs targeting PhCMoV are shown in Fig. S1. Multiple alignments of the available whole genome sequences of both viruses on NCBI showed high conservation among the different isolates at the locations targeted by these TomiRNAs (Table S2).
| Virus | Region | Prediction tools | ||||
|---|---|---|---|---|---|---|
| miRanda | RNAhybrid | RNA22 | Tapirhybrid | psRNATarget | ||
| 3′ | 0 | 0 | 5 | 1 | 4 | |
| N | 9 | 13 | 16 | 1 | 25 | |
| X | 0 | 1 | 0 | 0 | 3 | |
| P | 3 | 6 | 18 | 4 | 13 | |
| Y | 3 | 9 | 11 | 5 | 18 | |
| PhCMoV | M | 2 | 13 | 8 | 2 | 19 |
| G | 1 | 13 | 21 | 7 | 34 | |
| L | 30 | 86 | 87 | 24 | 92 | |
| 5′ | 1 | 2 | 3 | 1 | 2 | |
| UTRsa | 0 | 2 | 1 | 1 | 16 | |
| ToBRFV | 5′ | 0 | 0 | 0 | 0 | 2 |
| R (LC) | 13 | 121 | 34 | 28 | 84 | |
| R (SC) | 9 | 96 | 20 | 20 | 58 | |
| MP | 2 | 12 | 8 | 2 | 16 | |
| CP | 0 | 4 | 6 | 0 | 6 | |
| 3′ | 0 | 5 | 4 | 0 | 1 | |
| UTRs | 0 | 0 | 0 | 0 | 0 | |
Notes:
- N
-
nucleocapsid
- X
-
hypothetical protein with unknown function
- P
-
phosphoprotein
- Y
-
movement protein
- M
-
matrix protein
- G
-
glycoprotein and
- L
-
Large protein/RNA dependent RNA polymerase
- R (LC)
-
replicase large component subunit
- R (SC)
-
replicase small component
- MP
-
movement protein and
- CP
-
capsid protein
TomiRNAs’ target loci on tomato brown rugose fruit virus genome:
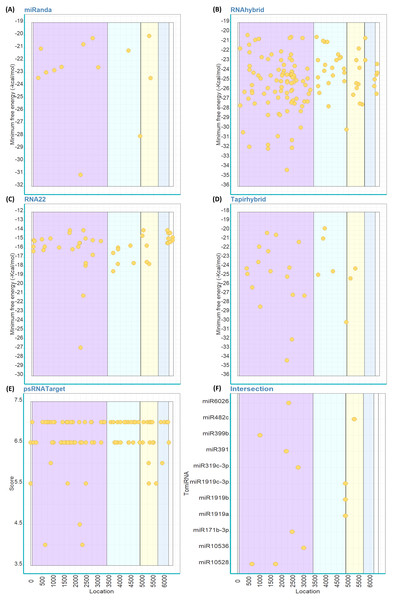
Out of the 147 mature TomiRNAs, miRanda predicted that 14 TomiRNAs are targeting 15 loci on ToBRFV genome (Fig. 3A). RNAhybrid predicted 142 TomiRNAs targeting 142 loci (Fig. 3B), RNA22: 41 TomiRNAs and 52 loci (Fig. 3C), Tapirhybrid: 27 TomiRNAs and 30 loci (Fig. 3D), and psRNATarget: 75 TomiRNAs and 109 loci (Fig. 3E). All the different regions of ToBRFV are predicted to be targets of TomiRNAs by at least one algorithm (Table 2).
Figure 3: Predicted target sites of tomato’s miRNAs on the genome of ToBRFV using miRanda (A), RNAhybrid (B), RNA22 (C), Tapirhybrid (D), psRNATarget (E) and (F) shows the common loci predicted by at least three miRNA target prediction algorithms.
The genes are highlighted as follow; R (LC) and R (SC) shared sequence [violet], rest of R (LC) (light blue), MP (yellow) and CP (blue); 5′and 3′ UTRs (white).Eleven TomiRNAs have common loci on the ToBRFV genome that were confirmed by three algorithms (Fig. 3F). Most of the predicted locations are on the shared nt sequence of the replicase genes R (LC) and R (SC) (Fig. 3F). These locations are targeted by miR10528 at loci (start positions 659 and 1734), miR399b (1024), miR391 (2213), miR6026 (2332), miR171-3p (2484), miR319c-3p (2755) and miR10536 (3009). miR1919a, b and c-3p are predicted to target the same position (starts at nt 4898) on the R (LC) gene sequence (Fig. 3F). The MP gene sequence is targeted by miR482c at position 5301 (Fig. 3F). For the CP gene, only one or two algorithms were able to predict TomiRNA target loci at all. The predicted folding structures of the precursor miRNAs targeting both viruses are shown in Fig. S2. Multiple alignments of the available whole genome sequences of ToBRFV on NCBI showed that the loci targeted by these TomiRNAs are highly conserved among the different isolates (Table S2).
About the best TomiRNA candidates targeting PhCMoV and ToBRFV genomes:
The 147 TomiRNAs used in this study can target different loci on the genomes of PhCMoV and ToBRFV as predicted by at least one algorithm. Of these loci, only 14 PhCMoV and 10 ToBRFV loci are supported by at least three algorithms, and have strong sequence complementarity, thus representing the best candidates. These candidate miRNAs were found to be involved in regulating plant genes expression and in biotic and abiotic stresses response.
miR164b-3p targets genes encoding stress-associated proteins and is suggested to be involved in the calcium ion homeostasis (Zhao et al., 2017). It was expressed in a drought-tolerant introgression line and was repressed by salt treatment (Liu et al., 2017; Xie et al., 2017; Zhao et al., 2017). Using RNA sequencing, miR166c-5p was significantly down-regulated in a drought-tolerant tomato introgression line, and in tomato leaf curl virus (ToLCV)-infected tomato (Liu et al., 2018; Tripathi et al., 2018). miR168a-3p regulates the expression of the ethylene receptor (Wang et al., 2017b; Wang et al., 2017a). miR168a-3p was repressed under drought stress, up-regulated in ToLCV-resistant tomato cv LA1777 and accumulated in potato virus Y (PVY)-infected tomato plants (Liu et al., 2018; Tripathi et al., 2018; Prigigallo et al., 2019). miR171b-3p was predicted to target SGN-E745132, an uncharacterized protein coding gene (Feng et al., 2014). In PVY-infected tomato plants, miR171b-3p was also accumulated and was not detected in healthy plants (Prigigallo et al., 2019).
miR319c-3p is involved in plant development and abiotic stress response (Shi et al., 2019). The expression of miR319c-3p decreased in response to heat stress, whereas its expression increased in moderately chill-tolerant and sensitive tomato genotypes (Shi et al., 2019). This suggested that it may have been responsible for the up-regulation of the tosinte branched/ cycloidea/ proliferating cell factors genes (TCP3, TCP29, and TCP2) (Shi et al., 2019). miR319c-3p specifically expressed in ToLCV-infected tomato “Pusa Ruby” but not in non-infected plants (Tripathi et al., 2018). The level of up-regulation was correlated with the infection and/or symptoms. However, TMV infection of tomato plants caused down-regulation of miR319c-3p (Abdelkhalek & Sanan-Mishra, 2019).
Microarray and northern hybridization showed a down-regulation of miR391 following ToLCNDV infection (Naqvi, Haq & Mukherjee, 2010). miR394-5p can target the LEAF CURLING RESPONSIVENESS (LCR) gene that is involved in the regulation of leaf, fruit and seed development; and its accumulation levels varied between the different tissue types of tomato plants (Tian et al., 2018). Its regulation affects the leaf curling phenotype (Song et al., 2012). miR396a-5p induces tomato disease susceptibility to Phytophthora infestans and Botrytis cinerea infections and enhances the tendency to produce reactive oxygen species (ROS) under pathogen-related biotic stress by suppressing target genes and upregulating salicylic acid (Chen et al., 2017). It was found that after drought stress, miR396a-5p was down-regulated in drought tolerant IL9–1 tomato, while it was up-regulated in the sensitive genotype M82 as determined by high-throughput sequencing (HTS) (Liu et al., 2017).
Functional analysis for tomato miRNAs targets revealed that miR408 targets copper-transporting ATPase PAA2 gene as a response to copper levels (Feng et al., 2014). miR408 was more abundant in leaves and closed flowers than in fruits (Moxon et al., 2008). miR477-3p targets transcription factor genes involved in plant development, and biotic and abiotic stress responses (Liu et al., 2018). It also targets resistance leucine-rich repeat receptor-like serine/threonine-protein kinase (RLK) (Hong et al., 2020). It was down-regulated under drought treatment and was not expressed in ToLCV-resistant tomato cv LA1777 (Liu et al., 2018; Tripathi et al., 2018).
Overexpression of miR482a transiently in Nicotiana benthamiana was associated with the decline in nucleotide-binding site leucine-rich repeat (NBS-LRR) mRNA (Eckardt, 2012). miR482a was up-regulated in tomato plants inoculated with the early blight causing fungus Alternaria solani and in ToLCNDV-infected plants (Pradhan et al., 2015; Sarkar et al., 2017). miR482c was also up-regulated in tomato plants infected with ToLCNDV and its overexpression induced enhanced susceptibility to late blight disease (Pradhan et al., 2015; Hong et al., 2019).
miR1916 is suggested to act as a negative regulator in the plant resistance to abiotic stress in Solanaceae (Chen, Meng & Luan, 2019). Overexpression of miR1916 in tomato reduced its drought tolerance, and its silence in transgenic plants increased drought stress resistance, significantly (Chen, Meng & Luan, 2019). It was down-regulated in tomato after P. infestans or B. cinerea infection (Chen et al., 2019). Its overexpression displayed significant enhancement in susceptibility to infection, as well as an increased tendency to ROS production. miR1918 was suggested to target the genome of ToLCV and to inhibit the viral replication (Naqvi et al., 2011). However, it enhanced tomato sensitivity to the infection of late blight disease causing pathogen “P. infestans” (Luan et al., 2016). miR1918 accumulated preferentially in the fruit (Moxon et al., 2008). Both miR1916 and miR1918 target tomato protein-expressing mRNAs (ESTs SGN-U322371 and SGN-U326398, respectively) (Moxon et al., 2008). miR1919a, miR1919b and miR1919c-3p are suggested to target the long non-coding RNA LncRNAZ114 associated with ethylene pathway in tomato (Wang et al., 2018a). The three TomiRNAs were up-regulated ToLCV-resistant tomato cv LA1777 (Tripathi et al., 2018). In addition, miR1919a was up-regulated under cold stress and down-regulated in a drought-sensitive genotype “M82”, whereas it was up-regulated in drought-tolerant genotype “IL9–1” (Chen et al., 2015; Liu et al., 2017).
miR5300 was predicted to target coiled coil-NBS-LRR domain genes involved in biotic stress response (Shivaprasad et al., 2012; Valiollahi et al., 2014; Pentimone et al., 2018). miR5300 was also found to be up-regulated in tomato roots inoculated with Pochonia chlamydosporia and down-regulated in ToLCNDV-infected plants (Pradhan et al., 2015; Pentimone et al., 2018). miR5303 was involved in growth-regulation, fruit development and ripening process in tomato (Mohorianu et al., 2011; Karlova et al., 2013; Yin et al., 2018; Zhao et al., 2018). miR10528, miR10536 and miR10541 were only identified in ToLCNDV-infected plants using HTS whereas miR399b appeared to be down-regulated in ToLCNDV-infected plants (Pradhan et al., 2015).
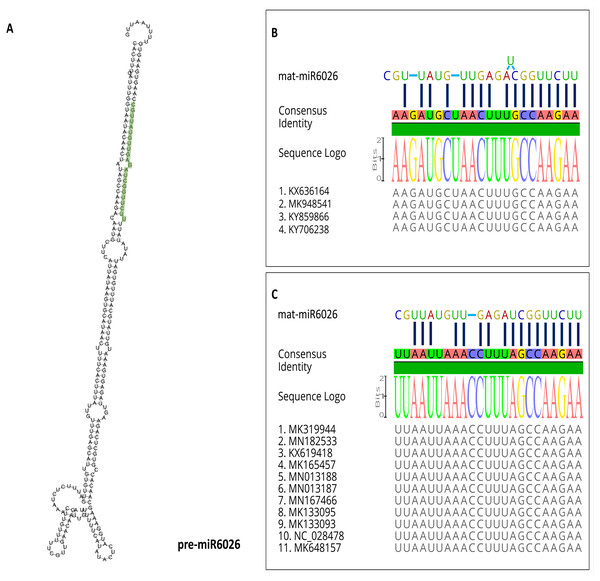
miR6026
miR6026 can target the genomes of PhCMoV and ToBRFV at the P gene of PhCMoV and the replicase components’ genes of ToBRFV (Figs. 2 and 3). Figure 4B shows the predicted folding structure of the precursor miR6026 (pre-miR6026). The sequence of the mature miR6026 (mat-miR6026) is UUC UUG GCU AGA GUU GUA UUG C (GenBank accession no. NR_108016). Nucleotide sequence alignments show that the sequences where the mat-miR6026 targets both PhCMoV and ToBRFV are conserved amongst all available isolates on NCBI (Figs. 4B and 4C). Seventeen nt out of 22 nt of mat-miR6026 can bind to both viral sequences (77% complementary) (Figs. 4B and 4C). The mat-miR6026 seed pairs with its match on the sequence of PhCMoV with 9mers and a supplementary of 8mers (Fig. 4B). With ToBRFV, the seed pairs with its match with 10mers and a supplementary of 7mers (Fig. 4C). The presence of conserved Watson–Crick pairing to the 5′ region of the miRNA “the miRNA seed”, reduces the occurrence of false-positive predictions (Lewis et al., 2003; Brennecke et al., 2005; Krek et al., 2005; Lewis, Burge & Bartel, 2005; Bartel, 2009). This perfect seed pairing also adds to the reliability of the prediction of the algorithms used (Lewis et al., 2003).
miR6026 is located on tomato chromosome 1 (position 832340 to 832581) (Li et al., 2012). It is predicted to regulate plant innate immune receptors (Li et al., 2012). It targets members of the DCL2 family, i.e., DCL2a, DCL2b, and DCL2d (Wang et al., 2018b). It has been demonstrated that miR6026 is also up-regulated in PVY-infected plants (Prigigallo et al., 2019). Moreover, miR6026 is also up-regulated in tomato roots inoculated with P. chlamydosporia endophytic hyphomycetes (Pentimone et al., 2018).
Figure 4: miR6026.
The predicted folding structure of the precursor miR6026 (pre-miR6026) with minimum free energy (A). The mature miR6026 (mat-miR6026) is highlighted in green. The predicted binding of the mat-miR6026 to the consensus sequence based on the alignment of the available PhCMoV (B) and ToBRFV (C) sequences on NCBI.These predicted miRNAs may be utilized to develop effective amiRNA constructs, which could be used to enhance the tomato plants immunity to both viruses. It is plausible that viral ORFs could be degraded after being recognized by amiRNA (Zhang et al., 2011; Song et al., 2014). Using these TomiRNA candidates for the transformation of tomato plants might not only defend plants against PhCMoV and ToBRFV but to other closely related viruses. Multiple amiRNAs can be inserted in a single gene expression cassette, which can be transformed to develop transgenic plant resistant to multiple viruses (Niu et al., 2006; Schwab et al., 2010). Therefore, future work will include the validation of these promising candidates in the development of PhCMoV- and ToBRFV-resistance in tomato plants.
Conclusion
In this study, a comprehensive computational approach was used to identify tomato-derived miRNAs for the silencing of PhCMoV and ToBRFV by RNA interference. Using five different bioinformatic tools with different algorithms, putative TomiRNAs targeting PhCMoV and ToBRFV have been predicted with high levels of conserved sites on the genomes of both viruses. Among the 14 best candidates of TomiRNAs targeting PhCMoV and the 11 targeting ToBRFV, miR6026 can target both viruses. The findings of this study may aid the development of PhCMoV- and ToBRFV-resistant tomato plants.
Supplemental Information
The precursor TomiRNAs share common targeting loci on PhCMoV and/or ToBRFV as predicted by at least three algorithms
List of TomiRNAs targeting PhCMoV and ToBRFV as predicted by miRanda, RNAhybrid, RNA22, Tapirhybrid and psRNATarget
The sequences of the mature TomiRNAs are listed and the numbers of loci targeted per TomiRNA are indicated.
The conserved loci on the consensus sequences of PhCMoV and ToBRFV targeted by TomiRNAs
The positions and percentages of loci conservation are indicated.
The predicted folding structure with minimum free energy of the precursor TomiRNAs targeting PhCMoV
The mature TomiRNAs are highlighted in green.
The predicted folding structure with minimum free energy of the precursor TomiRNAs targeting ToBRFV
The mature TomiRNAs are highlighted in green.