Interspecies interactions induce exploratory motility in Pseudomonas aeruginosa
Abstract
Microbes often live in multispecies communities where interactions among community members impact both the individual constituents and the surrounding environment. Here, we developed a system to visualize interspecies behaviors at initial encounters. By imaging two prevalent pathogens known to be coisolated from chronic illnesses, Pseudomonas aeruginosa and Staphylococcus aureus, we observed P. aeruginosa can modify surface motility in response to secreted factors from S. aureus. Upon sensing S. aureus, P. aeruginosa transitioned from collective to single-cell motility with an associated increase in speed and directedness – a behavior we refer to as ‘exploratory motility’. Explorer cells moved preferentially towards S. aureus and invaded S. aureus colonies through the action of the type IV pili. These studies reveal previously undescribed motility behaviors and lend insight into how P. aeruginosa senses and responds to other species. Identifying strategies to harness these interactions may open avenues for new antimicrobial strategies.
Introduction
While it is clear that many microbial infections do not occur with a single species, we have only recently begun to understand the profound impacts microbial species have on each other and patients (Nguyen and Oglesby-Sherrouse, 2016). Studies of dental biofilms, intestinal communities, chronic wounds, and respiratory infections in patients with cystic fibrosis (CF) demonstrate that community interactions influence microbial survival and disease progression (Limoli and Hoffman, 2019; Gabrilska and Rumbaugh, 2015). For example, we and others find an association between coisolation of Pseudomonas aeruginosa and Staphylococcus aureus from the CF airway or chronic wounds and poor patient outcomes, including decreased lung function and shortened life-spans (Limoli et al., 2016; Maliniak et al., 2016; Hubert et al., 2013). Laboratory studies also reveal interactions between these two pathogens can alter virulence factor production by one or both species, potentially influencing pathogenesis, persistence, and/or antibiotic susceptibility (Hotterbeekx et al., 2017). For example, in a model of coinfection on CF-derived bronchial epithelial cells, we observed P. aeruginosa and S. aureus form mixed microcolonies, which promotes the survival of S. aureus in the presence of vancomycin (Orazi and O'Toole, 2017). One strategy to improve outcomes for these coinfected patients may be to block harmful interspecies interactions before they begin.
Here, we designed a system to visualize early interactions between P. aeruginosa and S. aureus and follow single-cell behaviors over time with live-imaging. We show that P. aeruginosa can sense S. aureus secreted products from a distance and in turn, dramatically alter the motility behaviors of this Gram-negative bacterium. In response to S. aureus, individual P. aeruginosa cells transition from collective to single-cell movement, allowing exploration of the surrounding environment and directional movement towards S. aureus. We find that such ‘exploratory motility’ is driven primarily by the P. aeruginosa type IV pili (TFP) and modulated, in part, by the CheY-like response regulator, PilG. Importantly, we find P. aeruginosa is capable of responding to an array of bacterial species and strains recovered from a variety of sources, including CF patients, demonstrating a broad capacity for P. aeruginosa to sense other microbial species and modulate motility in response. Thus, we provide a new means to study polymicrobial interactions at the single-cell level and reveal that P. aeruginosa can sense the presence of other microbial species and dramatically, yet specifically, modify its behavior in response to such interspecies signals.
Results
S. aureus promotes exploratory motility in P. aeruginosa
To understand early microbial interactions, we established an in vitro coculture system to monitor P. aeruginosa and S. aureus at first encounters. Bacteria were inoculated at low cell densities between a coverslip and an agarose pad in minimal medium, supplemented with glucose and tryptone, and imaged with phase contrast time-lapse microscopy every 15 min for 8 hr. Alone, P. aeruginosa cells replicate and expand outward as raft-like groups, as previously described for P. aeruginosa surface-based motility (Anyan et al., 2014; Burrows, 2012) (Video 1; Figure 1, still montage, top row). In comparison, coincubation with S. aureus resulted in dramatically altered behavior (Video 2, Figure 1, still montage, bottom row). After two to three rounds of cell division, instead of remaining as a group, individual P. aeruginosa cells began to increase motility and moved as single-cells, suggesting that P. aeruginosa responds to the presence of S. aureus by altering motility behaviors. P. aeruginosa significantly inhibited S. aureus growth, as previously reported (see Figure 1—video 1 for representative time-lapse video of S. aureus alone).
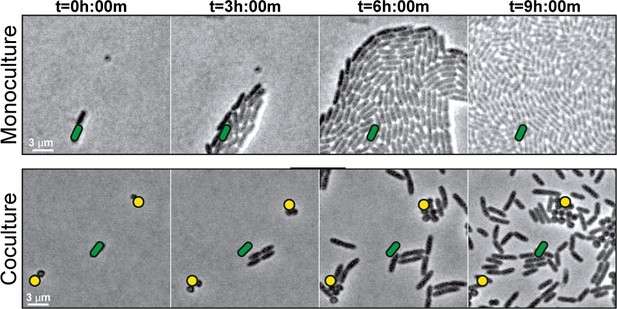
S. aureus increases P. aeruginosa motility.
Live-imaging of polymicrobial interactions. P. aeruginosa (rod-shaped) was inoculated between a coverslip and an agarose pad, either in monoculture (top) or in coculture with equal numbers of S. aureus (cocci-shaped, bottom). Images were acquired every 15 m for 9 hr. Representative snap-shots of Video 1 (top) and Video 2 (bottom) are shown. Founding cells identified in the first frame are indicated with green rods (P. aeruginosa) or yellow circles (S. aureus). The location of the founding cell is indicated in each subsequent frame for positional reference. At t = 9 h:00 m, the founding P. aeruginosa cell has moved outside the field of view.
WT P. aeruginosa in monoculture.
Duration 8 hr. Acquisition interval 15 m. Playback speed 3054x.
WT P. aeruginosa in coculture with WT S. aureus.
Duration 8 hr. Acquisition interval 15 m. Playback speed 3054x.
To visualize P. aeruginosa motility in the presence of S. aureus in more detail, the inoculating cell density was reduced (2–3 cells of each species per field of view), and images were taken at 5 s intervals for 8 hr. Video 3 shows images taken during hours 4–6 of coculture, when P. aeruginosa initiates single-cell movement under these conditions (Figure 2A, still montage). We observed a number of surprising behaviors by P. aeruginosa in the presence of S. aureus. P. aeruginosa cells initially replicated and remained in a raft (t = 4 h:28 m), as we and others have observed for P. aeruginosa in monoculture, but as the community approached S. aureus, individual cells: (1) exited the raft (t = 4 h:34 m), (2) moved with increased speed, and (3) moved towards and surrounded S. aureus, ‘explored’ the surface of the colony until (Figure 2A–B), (4) P. aeruginosa cells entered the S. aureus colony (Figure 2C, t = 6 hr), and finally, (5) by 8 hr, P. aeruginosa completely dismantled the S. aureus community (Figure 2D, t = 8 hr). P. aeruginosa was also observed to adopt a swift motion, beginning between 4.5–6 hr, moving in and out of the plane of focus during imaging (Video 4, yellow circle and Figure 2D, red arrows).
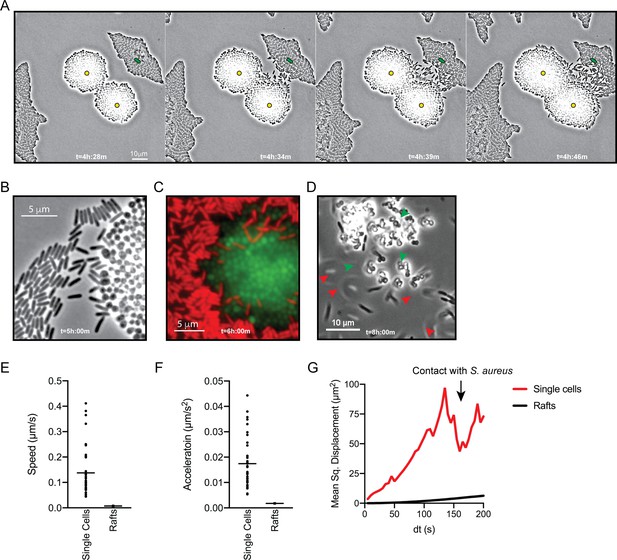
P. aeruginosa adopts an exploratory mode of motility in the presence of S. aureus.
Live-imaging of P. aeruginosa with WT S. aureus (Video 3). (A) Montage of representative snap-shots are shown beginning at 4 hr:28 m. Founding cells identified in the first frame are indicated with green rods (P. aeruginosa) or yellow circles (S. aureus). The location of the founding cell is indicated in each subsequent frame for positional reference. (B) Snap-shot at 5 hr, zoomed in to visualize single-cells. (C) Snap-shot at 6 hr of P. aeruginosa (mKO, red) and S. aureus (GFP, green) illustrating P. aeruginosa invasion into S. aureus colonies. (D) Snap-shot of coculture at 8 hr, showing disruption of S. aureus colonies (green arrows) and swift-moving P. aeruginosa cells out of the plane of focus (red arrows). Single P. aeruginosa cells and the leading edge of rafts were tracked in the presence of S. aureus and the speed (µm/s), acceleration (µm/s2), and mean squared displacement (µm2) for four independent videos are indicated, respectively, in (E – G).
WT P. aeruginosa in coculture with WT S. aureus.
Duration 10 m. 4 hr post inoculation. Acquisition interval 5 s. Playback speed 50x.
WT P. aeruginosa in coculture with WT S. aureus.
Duration 30 s. 4.5 hr post inoculation. Acquisition interval 50 ms. Playback speed 3x.
To quantitate the movement of P. aeruginosa single-cell motility in comparison to collective motility in rafts, the movement of individual cells and the leading edge of the rafts was tracked over time. In comparison to cells moving in rafts, individual cells moved with increased speed (µm/s), acceleration (µm/s2), and mean squared displacement (MSD, µm2) (Figure 2E, F and G, respectively). MSD represents a combined measure of both the speed and directional persistence of the cell, thus an increased MSD in single-cells over a change in time suggests single-cells exhibit directed motion, followed by a decreased MSD when P. aeruginosa cells reach the S. aureus colony.
P. aeruginosa type IV pili drive exploratory motility
How is P. aeruginosa motility generated in response to S. aureus? When grown in monoculture, P. aeruginosa performs cellular movement through the action of a single polar flagellum and the type IV pili (TFP) (Conrad et al., 2011; Gibiansky et al., 2010; Merritt et al., 2010). To determine how the observed P. aeruginosa ‘exploratory motility’ is generated, P. aeruginosa strains deficient in the production of either TFP (ΔpilA; encoding the pilin monomers), flagella (ΔflgK; encoding the flagella hook protein), or both (ΔpilA ΔflgK) were examined. Time-lapse images were taken as described for Figure 2, except they were acquired at 50 ms intervals for visualization of specific motility patterns. Representative videos and snap-shots (Figure 3A) were chosen at the time-point where P. aeruginosa was found to exhibit both slow and swift single-cell movements. The ΔpilA mutant was unable to move away from the group as single-cells (Video 5), as seen for the parental P. aeruginosa (Video 4), suggesting the TFP are required for P. aeruginosa exploratory motility. However, the swift movement observed in the WT (Figure 3A, see boxed inset with red arrows), was maintained in ΔpilA, demonstrating that TFP are not required for this behavior.
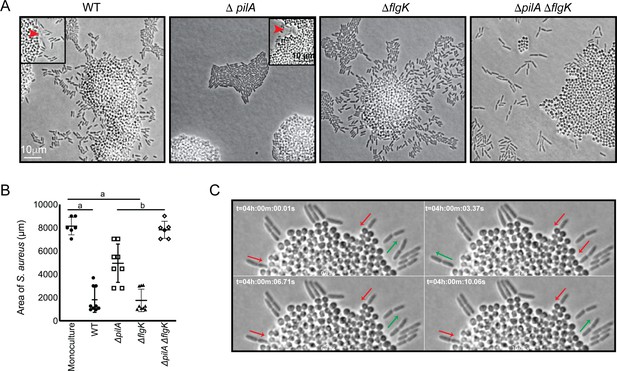
P. aeruginosa exploratory motility is driven by type IV pili.
Live-imaging of P. aeruginosa with WT S. aureus. (A) Representative snap-shots of coculture with WT S. aureus and P. aeruginosa (WT, ΔpilA, ΔflgK, and ΔpilA ΔflgK, left to right, Videos 4 and 5 and Figure 3—videos 1 and 2 respectively) are shown at t = 4.5 hr. Boxed insets show swift-moving P. aeruginosa cells out of the plane of focus (red arrows). (B) The area of S. aureus per frame in monoculture or in the presence of the indicated P. aeruginosa strain was calculated at t = 5 hr by dividing the total area occupied by S. aureus in a single frame by the number of S. aureus colonies. A minimum of four videos were analyzed per condition. The mean and standard deviation are indicated. Statistical significance was determined by one-way ANOVA followed by Tukey’s Multiple Comparisons Test - a indicates a statistically significant difference (p≤0.05) between S. aureus in monoculture and in the presence of either WT P. aeruginosa or ΔflgK; b indicates a statistically significant difference between ΔpilA and ΔpilA ΔflgK. (C) Representative snap-shots of Video 4, WT P. aeruginosa and S. aureus beginning at 4 hr with 50 ms intervals, showing back-and-forth motion. Red arrows indicate when a P. aeruginosa cell is moving in towards the S. aureus colony and green arrows indicate when a cell is moving away.
P. aeruginosa ΔpilA in coculture with WT S. aureus.
Duration 30 s. 4.5 hr post inoculation. Acquisition interval 50 ms. Playback speed 3x.
We next examined the ΔflgK mutant (Figure 3—video 1), which was seen to adopt the single-cell behaviors of the WT, except the swift movements were not observed, supporting the hypothesis that this movement is generated by the flagellum. These data suggest that while S. aureus modulates both TFP and flagella-mediated motility, the early events necessary for exploration (initiation of single-cell movement and directional movement towards S. aureus) require the TFP.
We also examined the response of a double ΔpilA ΔflgK mutant in the presence of S. aureus (Figure 3—video 2). Surprisingly, this mutant exhibited a phenotype distinct from either the WT, or the individual ΔpilA and ΔflgK mutants. P. aeruginosa cells deficient in both TFP and flagella were not only unable to produce the respective movements characteristic of these motility motors, but also were unable to remain within a raft-like group (Figure 3A). This behavior was not dependent upon the presence of S. aureus, as a similar pattern was observed for ΔpilA ΔflgK when visualized in the absence of S. aureus (Figure 3—figure supplement 1).
The influence of P. aeruginosa exploratory motility on S. aureus growth was also examined by measuring the area of the S. aureus colonies at 4.5 hr post-inoculation. S. aureus colonies were significantly smaller in the presence of WT P. aeruginosa or the ΔflgK mutant in comparison to S. aureus grown in monoculture (Figure 3B). However, in the presence the ΔpilA mutant, which was deficient in exploratory motility, colonies were significantly larger in comparison to WT and the ΔflgK mutant. Moreover, when grown in the presence of the double mutant, the area of the S. aureus colonies was not significantly different from S. aureus monoculture. These data suggest that ability of P. aeruginosa to perform exploratory motility influences P. aeruginosa inhibition of S. aureus growth.
While visualizing P. aeruginosa-S. aureus interactions at 50 ms intervals, we observed an additional P. aeruginosa behavior. When P. aeruginosa first encounters the S. aureus colony, the cell body orients perpendicular to the surface of the colony and moves back-and-forth (Video 4, blue circle and Figure 3C) and appears to move P. aeruginosa cells into the S. aureus colony. This behavior was only observed in the WT and ΔflgK mutant, suggesting that it is driven by the TFP.
Agr-regulated secreted S. aureus factors promote P. aeruginosa motility
Live-imaging suggests P. aeruginosa is capable of sensing S. aureus and initiating exploratory motility from a distance. Thus, we hypothesized that P. aeruginosa responds to S. aureus secreted factors. Since we observed that TFP were required for exploratory motility, we sought to develop a macroscopic assay where TFP motility could be monitored in the presence of S. aureus secreted factors only. Macroscopically, population-scale TFP-mediated twitching motility can be visualized by inoculating P. aeruginosa cells onto the bottom of a plate through 1.5% agar (sub-surface); cells move on the surface of the plate under the agar and can be stained with crystal violet for visualization (Turnbull and Whitchurch, 2014). To test the hypothesis that P. aeruginosa responds to S. aureus secreted products, cell-free supernatant derived from overnight cultures of WT S. aureus, (normalized to OD600 = 5.0) was spread on the plate, prior to pouring molten agar. S. aureus supernatant significantly increased the motility diameter of P. aeruginosa in a dose-dependent manner (Figure 4A), supporting the hypothesis that S. aureus secreted factors increase P. aeruginosa twitching motility.
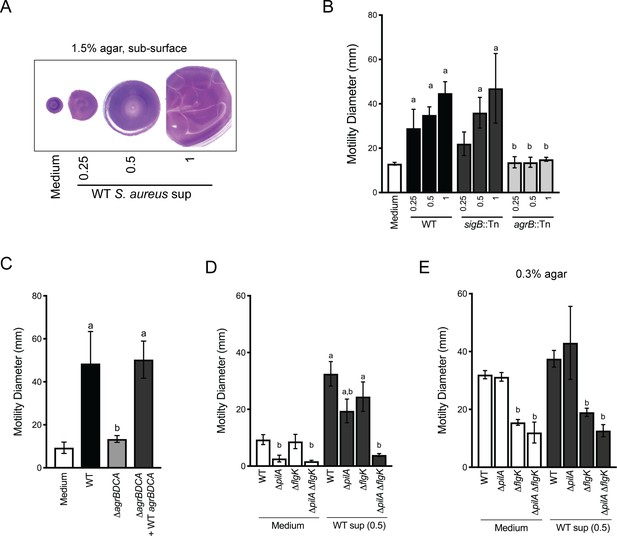
Agr-regulated secreted S. aureus factors increase P. aeruginosa motility.
(A – D) Motility of WT P. aeruginosa was monitored by macroscopic sub-surface inoculation assays in the presence of medium alone or cell-free supernatant from the indicated S. aureus strains. (A) illustrates representative motility zones stained with crystal violet for visualization. The dilution factor of the supernatant is indicated in A and B. Undiluted supernatant was used in C. In D and E, the motility of the indicated P. aeruginosa mutants was analyzed in the presence of medium alone or supernatant derived from WT S. aureus (0.5 dilution) under 1.5% agar (D) or within 0.3% agar (E). The mean and standard deviation are indicated for at least three biological replicates. Statistical significance was determined by one-way ANOVA followed by Tukey’s Multiple Comparisons Test - a indicates a statistically significant difference (p≤0.05) between the motility observed in the presence of S. aureus supernatant compared to medium alone, and b indicates a statistically significant difference (p≤0.05) between the motility observed in the mutant strain (S. aureus mutants in B and C, P. aeruginosa mutants in D and E) compared to the parental.
Two primary regulators of secreted factors in S. aureus are the alternative stress sigma factor, sigma B (SigB) (Bæk et al., 2013) and the accessory gene regulator (Agr) quorum sensing system (Nair et al., 2011). S. aureus strains with transposon insertions in either sigB or agrB (Fey et al., 2013) were examined for their ability to induce P. aeruginosa twitching motility (Figure 4B). While the sigB::Tn strain phenocopied the WT, the agrB::Tn mutant lost all ability to induce P. aeruginosa twitching motility, suggesting that Agr regulates the production of the factors promoting motility in P. aeruginosa. The agr operon is organized around two divergent promoters, P2 and P3, and generates two primary transcripts, RNAII and RNAIII, respectively. RNAII encodes AgrB, AgrD, AgrC, and AgrA (Le and Otto, 2015). To confirm a role for the Agr quorum sensing system, an unmarked deletion of agrBDCA was generated and complemented with WT agrBDCA. P. aeruginosa motility was examined in the presence of supernatant derived from these strains. As predicted, the ΔagrBDCA mutant was unable to enhance P. aeruginosa motility, and complementation restored activity to WT levels (Figure 4C).
To confirm that the TFP are necessary for increased P. aeruginosa motility in this assay, we examined the response of pilA-deficient P. aeruginosa to S. aureus supernatant. In the absence of S. aureus supernatant, the ΔpilA mutant was unable to perform twitching motility, as previously reported (Darzins, 1994). However, while motility was reduced in the presence of S. aureus supernatant, surprisingly, the ΔpilA mutant retained some ability to respond to S. aureus secreted factors (Figure 4D). Since we observed during live-imaging that S. aureus can increase P. aeruginosa flagella-mediated motility, in addition to TFP-mediated motility, we hypothesized that the response retained in the ΔpilA mutant was due to increased flagella-mediated motility. To test this hypothesis, we examined the response of ΔflgK and a double ΔpilA ΔflgK mutant to S. aureus supernatant. The single ΔflgK mutant phenotype trended lower, but was not significantly different from WT, while the motility of the ΔpilA ΔflgK mutant was reduced to levels not significantly different from ΔpilA or ΔpilA ΔflgK in the absence of supernatant (Figure 4D). These data support our observation from live-imaging that, while TFP were the primary contributors to increased motility, S. aureus secreted factors could increase both TFP- and flagella-mediated motility.
To formally examine the response of P. aeruginosa flagella-mediated motility to S. aureus supernatant, traditional macroscopic low-percentage agar assays that measure the contribution of flagella motility and chemotaxis were performed. Cell-free S. aureus supernatant was mixed into 0.3% agar prior to inoculating P. aeruginosa cells into the agar. The diameter of the P. aeruginosa motility zone showed a modest, but not significant increase in the presence of S. aureus supernatant, compared to medium alone (Figure 4E). To examine the role for flagella in response to S. aureus under swim assay conditions, the ΔflgK mutant was tested. As expected, flagella-mediated motility, both with and without S. aureus supernatant, was significantly reduced. To determine if TFP contribute under these assay conditions, the ΔpilA and ΔpilA ΔflgK mutants were also examined. The motility diameter of ΔpilA was not significantly different from WT, and the double mutant phenocopied the single ΔflgK mutant, suggesting that flagella are primarily responsible for the motility observed here.
P. aeruginosa biases the directionality of movement up a concentration gradient of S. aureus-secreted factors
Plate-based macroscopic motility assays measure both an absolute increase in motility and directional movement up a self-generated gradient (chemotaxis) as bacterial populations metabolize the available substrates and expand outward radially (Shapiro, 1984). While flagella-based movement through liquid has been extensively described in P. aeruginosa for a variety of chemoattractants, directional movement on a surface is poorly understood. Kearns et al. previously reported P. aeruginosa pili-mediated biased movement up a gradient of phosphatidylethanolamine (PE) on the surface of an agar plate (Kearns et al., 2001). To determine if P. aeruginosa is capable of moving up a previously established concentration gradient of S. aureus supernatant, supernatant derived from either WT or the ΔagrBDCA mutant of S. aureus was spotted onto the surface of 1.5% agar (containing buffered medium only, no carbon source). The secreted factors were allowed to diffuse and establish a gradient for approximately 24 hr, prior to inoculating P. aeruginosa onto the surface, as previously described (Kearns et al., 2001). Preferential movement of P. aeruginosa towards supernatant derived from WT S. aureus was observed, but not for medium alone or supernatant derived from the ΔagrBDCA mutant (Figure 5A). The response for the ΔpilA mutant was also examined and all motility was abrogated in this mutant, demonstrating motility observed in this assay is entirely pili-mediated. These data support the hypothesis that P. aeruginosa biases the directionality of TFP-mediated motility up a concentration gradient of S. aureus secreted factors.
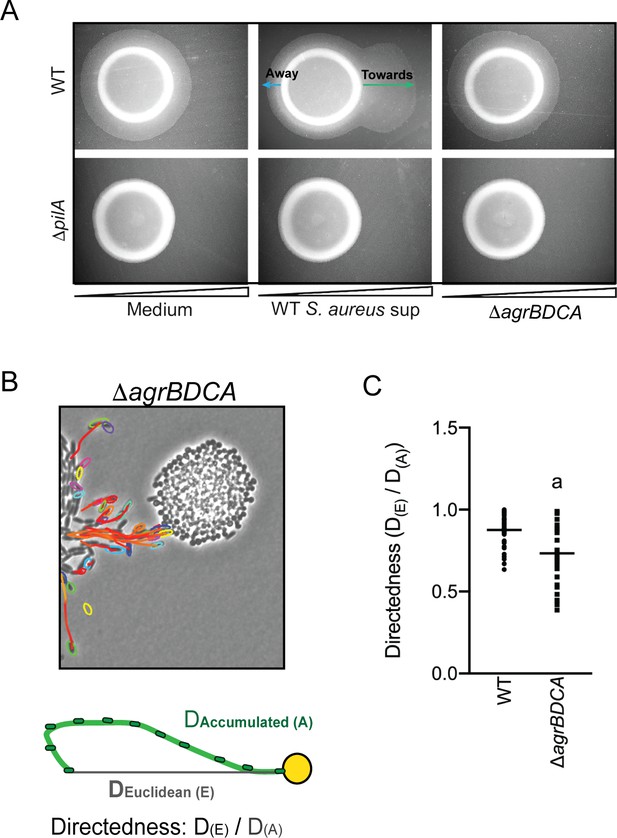
P. aeruginosa biases the directionality of movement up a concentration gradient of Agr-regulated secreted factors.
(A) A concentration gradient of either S. aureus growth medium (TSB), S. aureus supernatant derived from WT or ΔagrBDCA was established by spotting onto the surface of the agar and allowing a concentration gradient to establish by diffusion for approximately 24 hr, prior to spotting P. aeruginosa onto the agar (6 mm to the left) and surface-based motility imaged after 24 hr. Representative images of at least three independent experiments are shown. (B) Example of live-imaging of WT P. aeruginosa with S. aureus ΔagrBDCA with tracks of single-cells shown and schematic illustrating the methods for calculating the directedness. Single P. aeruginosa cells were tracked from first the frame a cell exited the raft to the frame where it first encounters S. aureus. The accumulated track distance, D(A), was measured for at least 30 cells in four independent videos and compared to the Euclidean distance, D(E), between the position of the cell in the first and last frame tracked. The ratio of D(E)/D(A) (Directedness) is shown in (C) for P. aeruginosa moving towards WT S. aureus compared to ΔagrBDCA with the mean indicated. Statistical significance was determined by an unpaired Student’s t-test (a = P ≤ 0.05).
We next asked if P. aeruginosa would also fail to migrate towards S. aureus deficient in Agr activity at the single-cell level. WT P. aeruginosa was visualized in coculture with ΔagrBDCA, as previously performed in the presence of WT S. aureus – with images acquired every 5 s for 8 hr (Video 3 for WT: Figure 2A and Video 6 for the ΔagrBDCA mutant: Figure 5B). In comparison to WT S. aureus, P. aeruginosa behavior was significantly altered in the presence of ΔagrBDCA. P. aeruginosa remained capable of initiating single-cell movement; however, once cells initiated single-cell movement, the path of their movement did not appear as directed towards the S. aureus colonies. In fact, some cells seemed to actively avoid the S. aureus colony all together.
WT P. aeruginosa in coculture with S. aureus ΔagrBDCA.
Duration 10 m. 4 hr post inoculation. Acquisition interval 5 s. Playback speed 50x.
To quantify the directedness of P. aeruginosa movement in the presence of WT and the ΔagrBDCA mutant, single P. aeruginosa cells were tracked from the first frame a cell exited the raft to the frame where it first encounters S. aureus (Figure 5B). The accumulated track distance, D(A), was measured and compared to the Euclidean distance, D(E), between the position of the cell in the first and last frame tracked. The directness of P. aeruginosa towards S. aureus was calculated as a ratio of D(E)/D(A). P. aeruginosa exhibited a significantly higher directedness ratio towards WT S. aureus compared to the ΔagrBDCA mutant (Figure 5C). These data support the hypothesis that Agr regulates the production of S. aureus secreted factors driving the directionality of P. aeruginosa motility.
The CheY-like response regulator, PilG, modulates P. aeruginosa response to S. aureus
How does P. aeruginosa sense the presence of S. aureus secreted products and initiate TFP-driven exploratory motility? P. aeruginosa encodes three known chemotaxis pathways: two flagella-mediated (che and che2) and a putative TFP-mediated system (pil-chp) (Darzins, 1994; Whitchurch et al., 2004). While several proteins encoded in the Pil-Chp pathway comprise a signal transduction pathway very similar (by gene homology) to the flagella-mediated chemotaxis pathway, their role in directional twitching motility remains unclear. A significant challenge lies in that many of these proteins are required for pilus assembly, thus teasing apart their requirement for motility per se versus regulation of a chemotactic response is challenging. Nonetheless, pilJ is predicted to encode the TFP methyl-accepting chemoreceptor protein (MCP) and by homology to the flagella MCPs, PilJ is expected to detect changes in the concentration of an attractant or repellent and initiate a signaling cascade to control twitching motility. To determine if PilJ is necessary for P. aeruginosa to sense S. aureus, we examined the response of a ΔpilJ mutant to S. aureus (WT) in the macroscopic sub-surface inoculation assay and by microscopic live-imaging. In the macroscopic assay, the ΔpilJ mutant exhibited low twitching motility, as previously described (Darzins, 1994) and responded to S. aureus supernatant to a similar degree as the ΔpilA mutant (Figure 6—figure supplement 1). However, examination by live-imaging revealed that ΔpilJ remained capable of performing surface motility and phenocopied the behavior of WT P. aeruginosa in the presence of S. aureus; moving as single-cells and with preferred directionality towards S. aureus (Figure 6A), suggesting that PilJ is not necessary for exploratory motility.
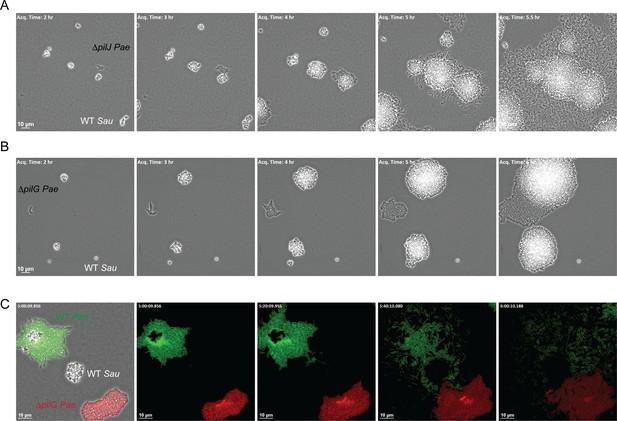
The CheY - like response regulator, PilG modulates P. aeruginosa response to S. aureus.
Representative snap-shots of P. aeruginosa ΔpilJ (A) and ΔpilG (B) in coculture with WT S. aureus. In (C), representative snap-shots of WT P. aeruginosa (GFP, green) with P. aeruginosa ΔpilG (mKate, red) and WT S. aureus (unmarked), visible only in the first frame (left) with phase contrast overlay.
A second protein encoded within the Pil-Chp chemosensory system that has been implicated in TFP-mediated chemotaxis is the CheY-like response regulator PilG, which regulates the ATPase necessary for TFP extension (PilB). Prior work demonstrates that while a ΔpilG mutant is non-motile in a macroscopic sub-surface inoculation assay (as previously described, Bertrand et al., 2010), in a microfluidic assay, individual surface attached P. aeruginosa cells were capable of performing twitching motility on a single-cell level; yet, they were unable to respond to a chemotactic gradient (Oliveira et al., 2016). We therefore examined a role for PilG in P. aeruginosa response to S. aureus. Similar to ΔpilJ we found that ΔpilG phenocopied the ΔpilA mutant in the macroscopic assay and was unable to generate twitching motility (Figure 6—figure supplement 1). However, during live-imaging, in comparison to ΔpilJ, ΔpilG was diminished in its capacity to increase motility in response to S. aureus (Figure 6B for PA14 and Figure 6—figure supplement 2 for PAO1, see discussion). Interestingly, we did note that in a few circumstances (3 of 9 videos), when a sufficient number of S. aureus cells were present (five or more founding cells per field of view) ΔpilG would eventually respond to the presence of S. aureus (Figure 6—figure supplement 3).
To confirm that ΔpilG exhibits a diminished response to S. aureus in comparison to WT, we imaged both WT P. aeruginosa and the isogeneic ΔpilG mutant in coculture with S. aureus simultaneously to account for variability in the number of S. aureus cells in the initial inoculum. P. aeruginosa strains were engineered to differentially express GFP (WT) or mKate (red, ΔpilG). Here the ΔpilG mutant displayed a diminished response to S. aureus in comparison to WT P. aeruginosa (Figure 6C). These data support the previous observations suggesting that a ΔpilG mutant is capable of performing TFP-mediated motility, but has a diminished response to chemotactic signals (Oliveira et al., 2016).
P. aeruginosa responds to a broad range of model organisms and CF pathogens
Our data thus far demonstrate that P. aeruginosa can sense S. aureus from a distance and modulate both flagella and TFP-mediated motility. To begin to examine if these behaviors might be relevant during airway infection in CF patients, we examined the capacity of a panel of P. aeruginosa isolates from CF patients to respond to S. aureus. Phenotypic loss of twitching motility in monoculture is often observed in chronic P. aeruginosa isolates from CF patients (Mayer-Hamblett et al., 2014), thus it was not surprising that two of the four isolates examined exhibited no detectable twitching motility in the absence of S. aureus. Nonetheless, each P. aeruginosa isolate was able to respond to S. aureus supernatant, although to varying degrees (Figure 7A). Importantly, even mucoid P. aeruginosa isolates (CFPBPA38, 43, 37), exhibited at least a two-fold increase in motility in the presence of S. aureus supernatant, above medium alone.
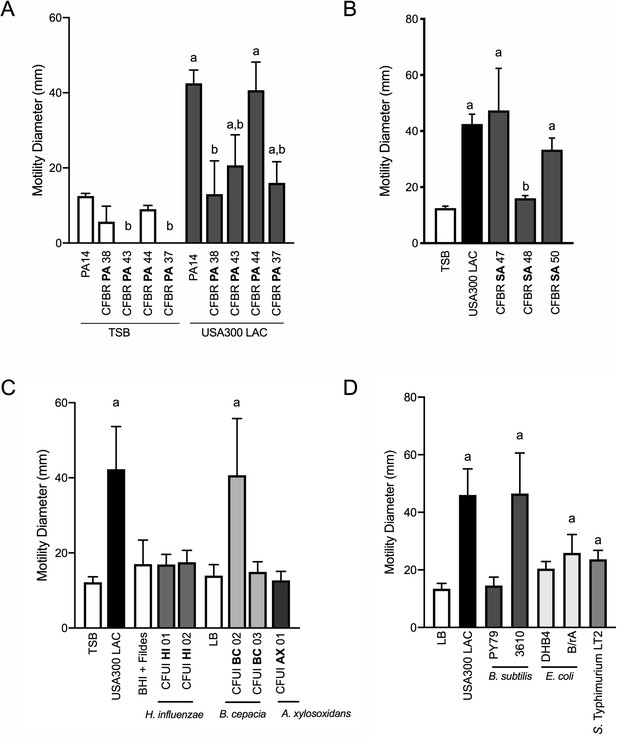
P. aeruginosa modulates motility in response to a range of CF clinical and non-clinical bacterial species.
The motility of clinical P. aeruginosa isolates (CFBR PA 38, 43, 44, and 37), in the presence of cell-free supernatant derived from S. aureus USA300 LAC (0.5 dilution) is indicated, in comparison to P. aeruginosa PA14 (A). The motility of P. aeruginosa laboratory strain PA14 was monitored in the presence of undiluted cell-free supernatant from clinical S. aureus CF isolates, CFBR SA 47, 48, and 50 (B), CF clinical isolates: H. influenzae, B. cepacia, and A. xylosoxidans (C), and non-CF species: E. coli, B. subtilis, and S. Typhimurium (D). Growth medium alone for each species was used as a negative control (TSB: S. aureus, BHI + Fildes: H. influenzae, LB: B. cepacia, A. xylosoxidans, B. subtilis, E. coli, and S. Typhimurium). The mean and standard deviation are indicated for at least three biological replicates. Statistical significance was determined by one-way ANOVA followed by Tukey’s Multiple Comparisons Test – a indicates a statistically significant difference (p≤0.05) between the motility observed in the presence of S. aureus supernatant compared to medium alone (A–D), and b indicates a statistically significant difference (p≤0.05) between the motility observed in the CF isolates, in comparison to laboratory strains (P. aeruginosa laboratory strain PA14 in (A) and S. aureus USA300 LAC in (B)).
Next, we asked if S. aureus isolates from CF patients are capable of inducing P. aeruginosa motility in the macroscopic twitching assay. Supernatant derived from two out of three clinical CF S. aureus isolates (each from different patients) was capable of inducing motility of the WT P. aeruginosa laboratory strain, to an extent not significantly different from that previously observed with WT S. aureus (Figure 7B). Since we previously observed Agr was necessary for S. aureus to promote motility, we hypothesized that CFBRSA48 was unable to induce P. aeruginosa due to reduced activity of the Agr quorum sensing system – an adaptation previously reported for S. aureus CF isolates (Nair et al., 2011; Goerke and Wolz, 2010). Since Agr also positively regulates the production of S. aureus β-hemolysin, hemolysis on sheep blood agar plates is often used as an indicator of Agr activity (Peng et al., 1988). However, all three clinical isolates maintain WT levels of hemolysis, suggesting Agr is active in these strains and an unknown mechanism accounts for reduced activity towards S. aureus in CFBRSA isolate 48 (Figure 7—figure supplement 1).
Can P. aeruginosa also respond to other CF pathogens, or is this behavior specific to S. aureus? To investigate this question, we examined clinical isolates from CF patients of three additional CF pathogens: Haemophilus influenzae, Burkholderia cepacia, and Achromobacter xylosoxidans. When possible, we examined multiple isolates for the ability of cell-free supernatant to enhance twitching motility in P. aeruginosa. We observed that one of two B. cepacia isolates tested enhanced P. aeruginosa motility to a similar degree as S. aureus, while none of those tested from H. influenzae or A. xylosoxidans significantly increased motility under these conditions (Figure 7C).
We then asked if P. aeruginosa is able to sense and respond more broadly to the presence of other bacterial species. Here we tested three additional non-CF organisms: Bacillus subtilis, Escherichia coli, and Salmonella Typhimurium. While the amount of P. aeruginosa motility observed varied between species and laboratory strains utilized, we observed a significant increase in P. aeruginosa motility in response to each organism tested, demonstrating that P. aeruginosa responds to a variety of bacterial species (Figure 7D).
To gain insight into the secreted microbial factors that promote P. aeruginosa motility, we performed preliminary biochemical analysis on S. aureus supernatant. First, cell-free supernatant was subjected to heat treatment at 95°C for 30 min. Heat treatment did not influence supernatant motility-inducing activity. However, protease treatment of the supernatant, with either proteinase k or trypsin, almost entirely eliminated activity (Figure 8A). These data suggest that the active factors are heat-stable proteins. Supporting this conclusion, we also found that the active factors were soluble in the aqueous fraction following hydrophobic extraction (Figure 8B). These data suggest the S. aureus factors responsible for promoting P. aeruginosa surface motility are proteinaceous in nature, but the activity is insensitive to denaturation by heat.
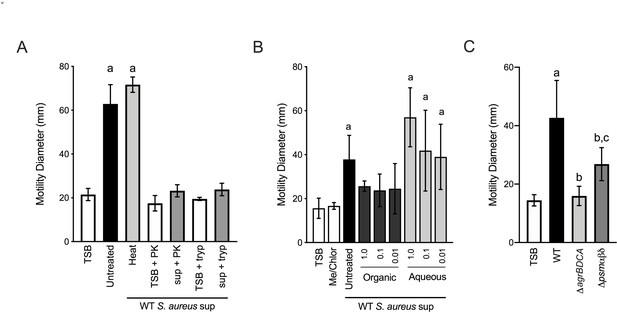
Phenol soluble modulins contribute to S. aureus-induced P. aeruginosa motility.
Motility of WT P. aeruginosa was monitored by macroscopic sub-surface inoculation assays in the presence of medium alone or cell-free supernatant derived from WT S. aureus treated with either heat, proteinase K (PK), or trypsin (tryp) in (A), methanol/chloroform extraction of the supernatant and subsequent analysis of the organic and aqueous fraction (methanol-chloroform (2:1, Me/Chlor) was included as the vehicle control) (B), or in the presence of untreated supernatant derived from WT or the indicated S. aureus mutants (0.5 dilution) in (C). The mean and standard deviation are indicated for at least three biological replicates. Statistical significance was determined by one-way ANOVA followed by Tukey’s Multiple Comparisons Test - a indicates a statistically significant difference (p≤0.05) between the motility observed in the presence of S. aureus supernatant compared to medium alone, b between the motility observed in the mutant strains compared to the parental, and c between the ΔagrBDCA and Δpsmαβδ mutants.
Together our biochemical and genetic evidence predict that heat-stable, Agr-regulated proteins and/or peptides promote P. aeruginosa motility. S. aureus produces such peptides referred to as phenol-soluble modulins (PSM). PSMs are amphipathic, multifunctional peptides that lyse many human cell types, are pro-inflammatory and chemoattractant to neutrophils, and influence S. aureus biofilm morphology and dispersal (Cheung et al., 2014). S. aureus produces five α-peptides (PSMα1–4 and δ-toxin) and two β-peptides (PSMβ1 and 2). PSMα are encoded by the psmα operon, PSMβ by the psmβ operon, and the δ-toxin (also called δ-hemolysin, Hld) is encoded within the RNAIII, regulatory RNA encoded from the agr operon. To determine if S. aureus PSMs might influence P. aeruginosa twitching motility, we collected supernatant from S. aureus deficient in the production of all seven PSMs, Δpsmα1–4 Δpsmβ1–2 and δ-toxin start (ATG) to stop (ATT) mutation, to preserve expression of RNAIII (referred to as Δpsmαβδ, from this point forward) (Syed et al., 2015). Supernatant derived from the Δpsmαβδ mutant showed a significantly reduced ability to promote P. aeruginosa motility in comparison to WT; however, the activity remained higher than ΔagrBDCA, suggesting one or more additional unidentified S. aureus factors also promote P. aeruginosa motility (Figure 8C).
Discussion
Current massive sequencing efforts yield unprecedented information regarding microbial community composition during infection, but fail to provide the necessary information required to functionally understand microbial behaviors in mixed species communities. Moreover, while the bulk assays most frequently utilized to study microbial interactions have provided insight into how bacterial species can influence community survival, metabolism, or virulence factor production (Hotterbeekx et al., 2017), we lack detailed information regarding how single-cells respond to the presence of another species at initial encounters. Here, we developed quantitative live-imaging methods to visualize the behaviors of two clinically important organisms and track their behavior over time. Through these studies, we uncovered microbial behaviors that could not have been predicted from bulk assays alone. We observed that P. aeruginosa is able to alter its behavior in the presence of S. aureus from a distance by responding to secreted factors. P. aeruginosa is then able to tune its motility patterns, adopting a behavior we refer to as ‘exploratory motility’.
What interspecies factors do P. aeruginosa sense and respond to? Our data thus far reveal that P. aeruginosa can not only respond to secreted products from S. aureus, but also more broadly to a range of model bacteria and CF pathogens, both Gram-positive and negative. P. aeruginosa increased surface motility to varying extents depending on the species and strain utilized – a feature we predict will inform the nature and breadth of factors to which P. aeruginosa is capable of responding. For S. aureus, we determined the factors are heat stable proteins regulated by the Agr quorum sensing system. This insight led to the determination that S. aureus PSMs contribute to the ability of S. aureus to induce surface motility in P. aeruginosa, but do not fully account for the observed phenotype, suggesting that additional unidentified factors are necessary. Moreover, S. aureus may also produce Agr-independent factors that influence P. aeruginosa motility, given that an Agr-deficient strain is unable to induce motility in the macroscopic sub-surface inoculation motility assay or directional motility on either the macroscopic or single-cell level, but remains capable of increasing single-cell motility during live-imaging. Thus, it is formally possible that separate factors are required for initiating single-cell motility and directional motion. Live-imaging also suggests that P. aeruginosa cells may actively avoid Agr-deficient S. aureus, raising the possibility that the Agr system negatively regulates a secreted factor that is unfavorable for P. aeruginosa.
How do PSMs promote P. aeruginosa motility? PSMs are amphipathic peptides that are able to reduce interfacial tension (IFT), that is, the cohesive or excess energy between molecules at an interface. While such surfactants have been extensively characterized to play an essential role in flagella-mediated bacterial surface motility, referred to as swarming (Kearns, 2010), their role in TFP-mediated motility is not well described. TFP generate bacterial movement on surfaces though polymerization and extension of pilus fibers, followed by pilus retraction, which pulls the cell body forward. Reduced IFT between the cell body and the surface, without affecting interaction between the pilus tip and the surface, may reduce drag on the cell body and reduce the force necessary to retract the pilus. TFP-mediated motility is most often described as a behavior where cells are primarily motile only in large groups. The behaviors for WT P. aeruginosa in monoculture observed here are consistent with previous descriptions of twitching motility, including the movement of cells in rafts, preferentially aligned along their long axis, and group tendril formation at the edges of the expanding community (Burrows, 2012). In contrast, in the presence of S. aureus, P. aeruginosa cells were seen to exit an expanding raft and transition to single-cell motility. Perhaps PSMs also reduce the IFT between neighboring P. aeruginosa cells, disrupting collective movement within rafts.
PSMs are known to promote S. aureus spreading on wet surfaces, in particular PSMα3 and δ-toxin, which possess high surfactant activity and low hydrophobicity (Tsompanidou et al., 2013). In the current study, we examined a mutant deficient in all seven PSMs, thus it is unknown if each S. aureus PSM possesses similar ability to promote motility and/or if their mechanism of action toward P. aeruginosa will also depend on their specific surfactant activity. Motility experiments in the presence of B. subtilis support the hypothesis that surfactants may increase TFP-mediated motility in P. aeruginosa. Two strains were examined, PY79 and 3610. 3610 induced P. aeruginosa motility similar to S. aureus. In comparison to PY79, which did not induce motility and is deficient in surfactant production, 3610 is an undomesticated strain that secretes a potent lipopeptide surfactant, called surfactin (Kearns and Losick, 2003). Thus, it is interesting to speculate that the different levels of surfactin account for the differential ability of these B. subtilis strains to promote P. aeruginosa motility.
PSMs can also form small transient pores in lipid membranes resulting in cell lysis of eukaryotic and some prokaryotic cells (Cheung et al., 2014). Thus, it is possible that interactions of PSMs with the P. aeruginosa outer membrane influence P. aeruginosa surface motility, independent of reduced IFT at the surface. For example, surfactin has been shown to influence B. subtilis quorum sensing and biofilm formation through the formation of membrane pores, resulting in potassium leakage. The resulting low cellular potassium activates the protein kinase, KinC, which regulates biofilm formation (López et al., 2009). Future studies will investigate if PSMs influence P. aeruginosa surface motility by reduction of IFT, interactions with the P. aeruginosa outer membrane, and/or alternative mechanism(s). Future studies are also necessary to determine if PSMs are sufficient to promote directional P. aeruginosa motility or if PSMs function in concert with additional S. aureus factors to increase surface motility per se in order to facilitate movement towards an additional stimulus.
While flagella-mediated chemotaxis has been extensively studied in P. aeruginosa, little is understood regarding how bacteria direct movement on a surface utilizing the TFP. While the P. aeruginosa Pil-Chp system harbors similarity to the well-studied Che system in E. coli, initial studies uncover significant differences in the regulation of this pathway. PilJ is a predicted chemoreceptor, by sequence similarity to the flagella chemotaxis system (Darzins, 1994); however, the functional role in chemotaxis remains unclear. In initial studies of TFP-mediated chemotaxis, P. aeruginosa was shown to direct motility on agar surfaces up a gradient of phosphatidylethanolamine (PE) (Kearns et al., 2001). While a ΔpilJ mutant did not exhibit preferential movement up a PE gradient, the lack of twitching motility in this mutant prohibited interpretation of a role for PilJ. Here, despite the lack of development of a macroscopic twitching motility zone, individual cells were capable of initiating motility in the presence of S. aureus when monitored by live-imaging, suggesting that P. aeruginosa is capable of sensing S. aureus and performing twitching motility in the absence of functional PilJ.
How does P. aeruginosa sense an external stimulus and transmit this into a mechanical response driving twitching motility? Twitching cells move by pulling themselves along by pili clustered at one pole. Cells can reverse direction by extending the pili from the opposite pole, changing the direction of movement along the long axis. These reversals have recently been suggested to be important for P. aeruginosa to direct movement up a chemotactic gradient while on a surface (Oliveira et al., 2016). In a ΔpilG mutant (the response regulator for pilus extension), cells were observed to perform fewer reversals in comparison to WT in response to a chemotactic gradient and thus were unable to bias the directionality of their movement. Here we examined a role for PilG in modulating a response to secreted factors from S. aureus and found a ΔpilG mutant was significantly diminished in the ability to respond. Both ΔpilJ and ΔpilG mutants have been previously reported to be deficient in producing traditional twitch rings in the macroscopic sub-surface inoculation assay (Luo et al., 2015; Bertrand et al., 2010; Darzins, 1994). Our observations here for PilJ and PilG, combined with those from Oliveira et al. suggest the collective movements of these mutants as measured in bulk assays may not be reflected on the single-cell level. However, in comparison to studies by Oliveira, we observed very little movement for P. aeruginosa in the absence of PilG. These studies utilized the P. aeruginosa parental strain PAO1 (in comparison to PA14 used here). We hypothesized that differences in parental strain background (PAO1 verses PA14) may account for the differences observed; however, we acquired the PAO1 parental and PAO1 ΔpilG mutant utilized in these studies and obtained similar results as observed for PA14 in coculture with S. aureus (Figure 6—figure supplement 2). Therefore, it is likely that experimental conditions influence to what extent P. aeruginosa is capable of performing TFP-mediated motility in the absence of PilG.
Myxococcus xanthus is one of the most notorious predatory bacteria – elaborating an array of social behaviors reminiscent of what we observe here for P. aeruginosa. M. xanthus coordinates a cooperative, density-dependent feeding behavior, resulting in propulsion of the cells rapidly through the colony of prey, leading to prey lysis, and nutrient acquisition. When M. xanthus contacts prey cells, pili-dependent reversals are stimulated, which keeps the cells ‘trapped’ near the prey colony, promoting contact-dependent prey killing and increased local concentration of antimicrobials (Muñoz-Dorado et al., 2016). Similarly, we observed that when P. aeruginosa encounters S. aureus, the P. aeruginosa cells appear to mount a coordinated response whereby P. aeruginosa surrounds the S. aureus colony and eventually invades the colony – a behavior referred to as the ‘wolf pack’ strategy. P. aeruginosa produces an arsenal of secreted antimicrobials shown to inhibit the growth of S. aureus, yet these secreted factors alone are insufficient for cellular lysis (Limoli et al., 2017). Thus, it is interesting to speculate that these behaviors function to synergistically increase cellular contacts necessary to kill S. aureus by a contact-dependent mechanism and/or to locally increase the concentration of secreted products.
By acquiring a fundamental understanding of how bacteria sense and respond to life with each other, we move closer to learning how to rationally manipulate interspecies behaviors during infection and in the environment. While for CF patients, this may mean preventing P. aeruginosa and S. aureus physical interactions, in other instances, we might bring species together who synergize to produce a beneficial compound.
Materials and methods
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Escherichia coli) | DH5α | Life Technologies | supE44 ΔlacU169 (ϕ80dlacZΔM15) hsdR17 thi-1 relA1 recA1 | |
| Strain, strain background (Escherichia coli) | S17 λpir | Life Technologies | TpR SmR recA, thi, pro, hsdR-M+RP4: 2-Tc:Mu:Km Tn7 λpir | |
| Strain, strain background (Escherichia coli) | B/rA | ATCC | ATCC 12407 | Obtained from David Weiss (Iowa) |
| Strain, strain background (Escherichia coli) | MC4100 | PMID: 11677609 | F− (araD139) Δ(lac)U169, strA, thi | Obtained from David Weiss (Iowa) |
| Strain, strain background (Pseudomonas aeruginosa) | PA14 (WT) | PMID: 7604262 | Non-mucoid prototroph SMC232(O’Toole Strain Collection) | |
| Strain, strain background (Pseudomonas aeruginosa) | PA14 mKO | PMID: 28084994 | attB:: PA1/04/03 -mKO Constitutively produces orange fluorescent protein | Obtained from Carey Nadell (Dartmouth) |
| Strain, strain background (Pseudomonas aeruginosa) | PA14 ΔpilA | PMID: 20233936 | SMC3782 (O’Toole StrainCollection) | |
| Strain, strain background (Pseudomonas aeruginosa) | PA14 ΔflgK | PMID: 28167523 | SMC5845 (O’Toole StrainCollection) | |
| Strain, strain background (Pseudomonas aeruginosa) | PA14 ΔpilA ΔflgK | PMID: 28167523 | SMC6595 (O’Toole StrainCollection) | |
| Strain, strain background (Pseudomonas aeruginosa) | PA14 ΔpilJ | PMID: 18178737 | SMC2992 (O’Toole Strain Collection) | |
| Strain, strain background (Pseudomonas aeruginosa) | PA14 ΔpilG | SMC4375 (O’Toole StrainCollection) | Obtained from Kyle Cady (Dartmouth) | |
| Strain, strain background (Pseudomonas aeruginosa) | PAO1 ΔpilG | PMID: 20008072 | Obtained from Joanne Engel (UCSF) | |
| Strain, strain background (Pseudomonas aeruginosa) | CFBR PA37 | PMID: 28325763 | Mucoid cystic fibrosis isolate | Obtained from the CF Biospecimen Registry (Emory) |
| Strain, strain background (Pseudomonas aeruginosa) | CFBR PA38 | PMID: 28325763 | Mucoid cysticfibrosis isolate | Obtained from the CF Biospecimen Registry (Emory) |
| Strain, strain background (Pseudomonas aeruginosa) | CFBR PA43 | PMID: 28325763 | Mucoid cysticfibrosis isolate | Obtained from the CF Biospecimen Registry (Emory) |
| Strain, strain background (Pseudomonas aeruginosa) | CFBR PA44 | PMID: 28325763 | Non-mucoid cystic fibrosis isolate | Obtained from the CF Biospecimen Registry (Emory) |
| Strain, strain background (Staphylococcus aureus) | USA300 LAC (WT) | PMID: 23404398 | USA300 CA-Methicillin resistant strain LACwithout plasmids | Obtained from Ambrose Cheung (Dartmouth) |
| Strain, strain background (Staphylococcus aureus) | USA300 LAC pCM29 | PMID: 20829608 | sarAP1-sGFP | Obtained from Kenneth Bayles (Nebraska) |
| Strain, strain background (Staphylococcus aureus) | USA300 LAC agrB::TnMar | PMID: 23404398 | Obtained from the Nebraska Transposon Mutant Library | |
| Strain, strain background (Staphylococcus aureus) | USA300 LACsigB::TnMar | PMID: 23404398 | Obtained from the Nebraska Transposon Mutant Library | |
| Strain, strain background (Staphylococcus aureus) | USA300 LACΔagrBDCA | This study | See Materials and methods | |
| Strain, strain background (Staphylococcus aureus) | USA300 LAC ΔagrBDCA + WT agrBDCA | This study | See Materials and methods | |
| Strain, strain background (Staphylococcus aureus) | CFBR SA47 | PMID: 28325763 | Cystic fibrosis isolate | Obtained from the Nebraska Transposon Mutant Library |
| Strain, strain background (Staphylococcus aureus) | CFBR SA48 | PMID: 28325763 | Cystic fibrosis isolate | Obtained from the CF Biospecimen Registry (Emory) |
| Strain, strain background (Staphylococcus aureus) | CFBR SA50 | PMID: 28325763 | Cystic fibrosis isolate | Obtained from the CF Biospecimen Registry (Emory) |
| Strain, strain background (Haemophilus influenzae) | CFUI HI01 | This study | Cystic fibrosis isolate | Obtained from U Iowa CF Biobank |
| Strain, strain background (Haemophilus influenzae) | CFUI HI02 | This study | Cystic fibrosis isolate | Obtained from U Iowa CF Biobank |
| Strain, strain background (Burkholderia cepacia) | CFUI BC02 | This study | Cystic fibrosis isolate | Obtained from U Iowa CF Biobank |
| Strain, strain background (Burkholderia cepacia) | CFUI BC03 | This study | Cystic fibrosis isolate | Obtained from U Iowa CF Biobank |
| Strain, strain background (Achromobacter xylosoxidans) | CFUI AX01 | This study | Cystic fibrosis isolate | Obtained from U Iowa CF Biobank |
| Strain, strain background (Bacillus subtilis) | PY79 | PMID: 6093169 | Prototrophic derivative of B. subtilis 168 | Obtained from Craig Ellermeier (Iowa) |
| Strain, strain background (Bacillus subtilis) | 3610 | PMID: 11572999 | NCIB 3610:Marburg ‘wild’ isolate | Obtained from Craig Ellermeier (Iowa) |
| Strain, strain background (Salmonella enterica serovar Typhimurium) | LT2 | PMID: 11677609 | Obtained from Bradley Jones (Iowa) | |
| Strain, strain background (Staphylococcus aureus) | USA300 LACΔpsmαβδ | PMID: 26077761 | psmΔα1–4 Δβ1–2 δATG-ATT | Obtained from Daniel Wozniak (OSU) |
| Recombinant DNA reagent | pMAD | PMID: 15528558 | E. coli-S. aureus shuttle vector. OripE194TS ermC blaC bgaB | |
| Commercial assay or kit | Gibson Assembly Cloning Kit | New England BioLabs | E5510 | |
| Software, algorithm | NIS-Elements AR | Nikon | Version 5.2 |
Bacterial strains and culture conditions
Request a detailed protocolP. aeruginosa and E. coli were routinely cultured in lysogeny broth (LB; 1% tryptone, 0.5% yeast extract, 1% sodium chloride) and S. aureus in tryptic soy broth (TSB, Becton Dickenson) at 37°C, with aeration. For coculture assays, both species were grown in TSB or M8 minimal medium (48 mM sodium phosphate dibasic, 22 mM potassium phosphate monobasic, 8.6 mM sodium chloride, 2.0 mM magnesium sulfate, 0.1 mM calcium chloride) supplemented with 1% glucose and tryptone. When necessary for strain construction, medium was supplemented with the following antibiotics: gentamicin (10 µg/ml E. coli; 30 µg/ml P. aeruginosa), ampicillin (100 µg/ml E. coli), carbenicillin (200 µg/ml P. aeruginosa), or chloramphenicol (10 µg/ml S. aureus).
P. aeruginosa and S. aureus clinical isolates were obtained from the CF Biospecimen Registry (CFBR) at the Children’s Healthcare of Atlanta and Emory University CF Discovery Core and the B. cepacia, A. xylosoxidans, and H. influenzae isolates were obtained from the University of Iowa Cystic Fibrosis Biobank. B cepacia, A. xylosoxidans, B. subtilis, and S. Typhimurium were grown in LB and H. influenzae in Brain Heart Infusion (BHI) broth supplemented with 5% Fildes Enrichment at 37°C with aeration.
S. aureus genetic manipulation. In-frame deletion of the agrBDCA operon in a S. aureus USA300 LAC strain (JE2) was generated using pMAD-mediated allelic replacement (Arnaud et al., 2004). Briefly, a pMAD deletion vector was created by Gibson assembly (Gibson et al., 2009) using EcoRI/BamHI-linearized pMAD and PCR amplicons of 1 kb regions up- and downstream of agrBDCA (primer sets agr-a/agr-b and agr-c/agr-d, respectively, Table 1). The vector to chromosomally complement this deletion strain was similarly produced using Gibson assembly on the EcoRI/BamHI-lineararized pMAD vector and the PCR product of the agr-a/agr-d primers. Following the standard protocol of heat shift on selective media, strains with successful deletions and restorations of agrBDCA were verified by PCR analysis and chromosomal DNA sequencing.
Oligonucleotides used in this study.
| Name | Sequence | Reference |
|---|---|---|
| agr-a | CGCGGATCCTACATAGCACTGAGTCCAAGG | This study |
| agr-b | GCCGTTAACTGACTTTATTATCTTTTTTACACCACTCTCCTCACTG | This study |
| agr-c | CAGTGAGGAGAGTGGTGTAAAAAAGATAATAAAGTCAGTTAACGGC | This study |
| agr-d | CCGGAATTCCAGTTATTAGCAGGATTTTAGC | This study |
Live-imaging of interspecies interactions
Request a detailed protocolBacteria were inoculated between a coverslip and an agarose pad and imaged with time-lapse microscopy. Pads were made by adding 900 µl of M8 medium with 1% glucose, 1% tryptone, and 2% molten agarose to a 10 mm diameter silicone mold on a coverslip. Pads were allowed to dry for 1 hr at room temperature, followed by 1 hr at 37°C. Meanwhile, bacteria were prepared by growing to mid-log phase in M8 medium with 1% glucose and 1% tryptone, diluting to OD600 = 0.15 in warm media, and mixing P. aeruginosa and S. aureus 1:1. 2 µl of inoculum was spotted evenly onto the center of a warm 35 mm glass bottom dish, #1.5 mm coverglass (Cellvis). The agarose pad was removed from the mold and placed on top of the bacterial inoculum. Bacteria were immediately imaged with an inverted Nikon TiE or Ti2 at 37°C for 8 hr. Images were acquired with a 100x oil objective (1.45NA) with phase contrast and an ORCA Flash4.0 Digital CMOS camera (Hammamatsu).
Videos were generated and cells tracked and analyzed in Nikon Elements AR. For single-cells, binary images were generated and single P. aeruginosa cells were identified and tracking began when the first P. aeruginosa cell exited a raft. Rafts were tracked up until the time point where single-cell tracking began. Rafts edges were manually identified. The speed (µm/s), acceleration (µm/s2), mean squared displacement (µm2), accumulated track distance, D(A), and the Euclidean distance, D(E), were measured for at least 30 tracks (for single-cells) in four independent videos. The area of S. aureus was determined by dividing the total area occupied in a field of view divided by the number of colonies.
Macroscopic coculture assays
Request a detailed protocolP. aeruginosa sub-surface twitch (Turnbull and Whitchurch, 2014) and 0.3% soft agar motility assays (Ha et al., 2014) were performed as previously described with modifications for treatment with bacterial supernatant. Supernatant was derived by growing the indicated species overnight centrifuging to pellet cells, and supernatant filtered through a 0.22 µm filter. S. aureus was grown in TSB and strains normalized to OD600 = 5.0 (and subsequently diluted to the indicated concentrations). For soft agar assays, cell-free supernatant was mixed at the indicated concentrations with cooled, but still molten agar prior to pouring. For subsurface inoculation assays, 100 µl of supernatant was spread onto the bottom of a petri plate before pouring media (1.5% agar). P. aeruginosa was grown to OD600 = 2.0 in TSB and inoculated into motility plates by either stabbing with a toothpick halfway through the agar (0.3%), all the way though (sub-surface). Plates were incubated at 37°C for 24 hr, followed by 24 hr at room temperature. For twitch plates, agar was dropped out and the P. aeruginosa biomass stained with 1% crystal violet for visualization. The diameter of the motility zones was measured in mm.
Biochemical analysis of S. aureus supernatant
Request a detailed protocolSupernatant was collected as described above and exposed to the following conditions: 1. Heat: 95°C for 30 min. 2. Protease treatment: Proteinase K (400 µg/ml) at 55°C for 30 min or trypsin (2.5 µg/ml) at 37°C for four hours with gentle shaking, followed by heat inactivation at 95°C for 20 min. 3. Hydrophobic extraction: Performed according to the guidelines of Bligh and Dyer (1959). In brief, a volume of 3.75 ml of methanol-chloroform (2:1) was added to 1.6 ml of S. aureus supernatant and vortexed for 1 hr. The suspension was centrifuged at 10,000 x g for 5 min and the aqueous phase was saved. The organic phase was then reextracted with 4.75 ml of methanol-chloroform-water (2:1:0.8) and centrifuged again, and the extracted organic phases were combined.
Directional twitching motility assay
Request a detailed protocolExperiments were performed as previously described (Miller et al., 2008). In brief, buffered agar plates (10 mM Tris, pH 7.6; 8 mM MgSO4; 1 mM NaPO4, pH 7.6; and 1.5% agar) were poured and dried at room temperature for 24 hr. 4 µl of supernatant or medium (TSB) was spotted on the plates, and gradients were established by incubating the plates at 30°C for 24 hr. Supernatants were prepared as described above. P. aeruginosa strains were grown to early stationary phase, normalized to an OD600 of 1.2 and 2 mls pelleted and resuspended in 100 ml of MOPS buffer (10 mM MOPS, pH 7.6, and 8 mM MgSO4). 2 µl of this cell suspension was spotted approximately 6 mm from the center of the supernatant spot. Plates were incubated for 24 hr at 37°C followed by 24 hr at room temperature before images were taken of the twitching zones.
Data availability
All data generated or analyzed during this study are included in the manuscript and supporting files.
References
-
New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, Gram-positive bacteriaApplied and Environmental Microbiology 70:6887–6891.https://doi.org/10.1128/AEM.70.11.6887-6891.2004
-
A rapid method of total lipid extraction and purificationCanadian Journal of Biochemistry and Physiology 37:911–917.https://doi.org/10.1139/y59-099
-
Pseudomonas aeruginosa twitching motility: type IV pili in actionAnnual Review of Microbiology 66:493–520.https://doi.org/10.1146/annurev-micro-092611-150055
-
Phenol-soluble modulins--critical determinants of staphylococcal virulenceFEMS Microbiology Reviews 38:698–719.https://doi.org/10.1111/1574-6976.12057
-
Flagella and pili-mediated near-surface single-cell motility mechanisms in P. aeruginosaBiophysical Journal 100:1608–1616.https://doi.org/10.1016/j.bpj.2011.02.020
-
Characterization of a Pseudomonas aeruginosa gene cluster involved in pilus biosynthesis and twitching motility: sequence similarity to the chemotaxis proteins of enterics and the gliding bacterium Myxococcus xanthusMolecular Microbiology 11:137–153.https://doi.org/10.1111/j.1365-2958.1994.tb00296.x
-
Biofilm models of polymicrobial infectionFuture Microbiology 10:1997–2015.https://doi.org/10.2217/fmb.15.109
-
Adaptation of Staphylococcus aureus to the cystic fibrosis lungInternational Journal of Medical Microbiology 300:520–525.https://doi.org/10.1016/j.ijmm.2010.08.003
-
Plate-Based assay for swimming motility in Pseudomonas aeruginosaMethods in Molecular Biology 1149:59–65.https://doi.org/10.1007/978-1-4939-0473-0_7
-
In vivo and in vitro Interactions between Pseudomonas aeruginosa and Staphylococcus sppFrontiers in Cellular and Infection Microbiology 7:106.https://doi.org/10.3389/fcimb.2017.00106
-
A field guide to bacterial swarming motilityNature Reviews Microbiology 8:634–644.https://doi.org/10.1038/nrmicro2405
-
Swarming motility in undomesticated Bacillus subtilisMolecular Microbiology 49:581–590.https://doi.org/10.1046/j.1365-2958.2003.03584.x
-
Quorum-sensing regulation in staphylococci—an overviewFrontiers in Microbiology 6:1174.https://doi.org/10.3389/fmicb.2015.01174
-
Staphylococcus aureus and Pseudomonas aeruginosa co-infection is associated with cystic fibrosis-related diabetes and poor clinical outcomesEuropean Journal of Clinical Microbiology & Infectious Diseases 35:947–953.https://doi.org/10.1007/s10096-016-2621-0
-
Pseudomonas aeruginosa in vitro phenotypes distinguish cystic fibrosis infection stages and outcomesAmerican Journal of Respiratory and Critical Care Medicine 190:289–297.https://doi.org/10.1164/rccm.201404-0681OC
-
Myxobacteria: moving, killing, feeding, and surviving togetherFrontiers in Microbiology 7:e1004474.https://doi.org/10.3389/fmicb.2016.00781
-
Interactions between Pseudomonas aeruginosa and Staphylococcus aureus during co-cultivations and polymicrobial infectionsApplied Microbiology and Biotechnology 100:6141–6148.https://doi.org/10.1007/s00253-016-7596-3
-
Distinct roles of phenol-soluble modulins in spreading of Staphylococcus aureus on wet surfacesApplied and Environmental Microbiology 79:886–895.https://doi.org/10.1128/AEM.03157-12
-
Motility assay: twitching motilityMethods in Molecular Biology 1149:73–86.https://doi.org/10.1007/978-1-4939-0473-0_9
Article and author information
Author details
Funding
Cystic Fibrosis Foundation (Postdoctoral Fellowship LIMOLI15F0)
- Dominique H Limoli
Cystic Fibrosis Foundation (CFF Postdoc-to-Faculty Transition Award LIMOLI18F5)
- Dominique H Limoli
National Institutes of Health (Grant R37 AI83256)
- George O'Toole
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Acknowledgements
This work was supported by funding from NIH Grant R37 AI83256 (GAO), CFF Postdoctoral Fellowship LIMOLI15F0, and CFF Postdoc-to-Faculty Transition Award LIMOLI18F5. We thank Drs. Ethan Garner, Gerard Wong, Jeffrey Meisner, and Minsu Kim for consultation on imaging studies, Kenneth Bayles, David Weiss, Craig Ellermeier, Bradley Jones, and Joanne Engel for bacterial strains, and Timothy Yahr for thoughtful discussions and careful reading of the manuscript.
Version history
- Received: April 3, 2019
- Accepted: October 30, 2019
- Accepted Manuscript published: November 12, 2019 (version 1)
- Version of Record published: December 13, 2019 (version 2)
Copyright
© 2019, Limoli et al.
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 7,135
- Page views
-
- 1,043
- Downloads
-
- 42
- Citations
Article citation count generated by polling the highest count across the following sources: Scopus, Crossref, PubMed Central.
Download links
Downloads (link to download the article as PDF)
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools)
Further reading
-
- Microbiology and Infectious Disease
Neonatal meningitis is a devastating disease associated with high mortality and neurological sequelae. Escherichia coli is the second most common cause of neonatal meningitis in full-term infants (herein NMEC) and the most common cause of meningitis in preterm neonates. Here, we investigated the genomic relatedness of a collection of 58 NMEC isolates spanning 1974–2020 and isolated from seven different geographic regions. We show NMEC are comprised of diverse sequence types (STs), with ST95 (34.5%) and ST1193 (15.5%) the most common. No single virulence gene profile was conserved in all isolates; however, genes encoding fimbrial adhesins, iron acquisition systems, the K1 capsule, and O antigen types O18, O75, and O2 were most prevalent. Antibiotic resistance genes occurred infrequently in our collection. We also monitored the infection dynamics in three patients that suffered recrudescent invasive infection caused by the original infecting isolate despite appropriate antibiotic treatment based on antibiogram profile and resistance genotype. These patients exhibited severe gut dysbiosis. In one patient, the causative NMEC isolate was also detected in the fecal flora at the time of the second infection episode and after treatment. Thus, although antibiotics are the standard of care for NMEC treatment, our data suggest that failure to eliminate the causative NMEC that resides intestinally can lead to the existence of a refractory reservoir that may seed recrudescent infection.
-
- Microbiology and Infectious Disease
A productive HIV-1 infection in humans is often established by transmission and propagation of a single transmitted/founder (T/F) virus, which then evolves into a complex mixture of variants during the lifetime of infection. An effective HIV-1 vaccine should elicit broad immune responses in order to block the entry of diverse T/F viruses. Currently, no such vaccine exists. An in-depth study of escape variants emerging under host immune pressure during very early stages of infection might provide insights into such a HIV-1 vaccine design. Here, in a rare longitudinal study involving HIV-1 infected individuals just days after infection in the absence of antiretroviral therapy, we discovered a remarkable genetic shift that resulted in near complete disappearance of the original T/F virus and appearance of a variant with H173Y mutation in the variable V2 domain of the HIV-1 envelope protein. This coincided with the disappearance of the first wave of strictly H173-specific antibodies and emergence of a second wave of Y173-specific antibodies with increased breadth. Structural analyses indicated conformational dynamism of the envelope protein which likely allowed selection of escape variants with a conformational switch in the V2 domain from an α-helix (H173) to a β-strand (Y173) and induction of broadly reactive antibody responses. This differential breadth due to a single mutational change was also recapitulated in a mouse model. Rationally designed combinatorial libraries containing 54 conformational variants of V2 domain around position 173 further demonstrated increased breadth of antibody responses elicited to diverse HIV-1 envelope proteins. These results offer new insights into designing broadly effective HIV-1 vaccines.






